HTML
-
Studies of bacterial viruses, or bacteriophages, which were originally discovered in parallel (Duckworth, 1976) by F. H. d'Herelle (1917) and F. W. Twort (1915), have been fundamental to the development of a modern molecular understanding of biology. In spite of this, following the introduction of antibiotics, the possibility of using them as anti-infective agents became neglected almost immediately, worldwide. Recently, however, the introduction of new antibiotics into medical practice has seemed almost futile, owing to the emergence and widespread occurrence of multidrug-resistant pathogenic bacteria strains (Fowler et al., 2014). As a result, there has arisen a movement to return to the use of bacteriophages in general medical practice for the treatment of at least some bacterial infections (mainly superficial pyogenic infections). In addition, there are proposals to use phage-encoded enzymes in therapy (Rodríguez-Rubio et al., 2012; Schmelcher et al., 2012; Briers et al., 2014).
One problem about phage therapy is its uncertainty safety. Indeed, it has transpired that the development of methods for the rapid analysis of phage genomes has had virtually no influence on ou r understanding of the functions of most of the products encoded by phage genes. As an example, 157 open reading frames (ORFs) have been identified in the genome of Pseudomonas aeruginosa virulent phage PaP1 (Lu et al., 2013). Of these, although 143 encode homologs of proteins that have already been cited in protein databases, the precise functions are known for only 38 of them. Moreover, there remain 14 unique genes that encode gene products with completely unknown functions. The implications of this are that the events which occur in bacteria following infection with phages can be described only in general terms. Furthermore, it is sometimes the case that phages showing only minor differences between their genomes can nevertheless display differences that may affect the efficacy and safety of phage treatments (see later).
Most of the publications that emphasize the safety and high degree of efficacy of phage therapies, phage products and the uses of phages as antiseptics usually refer to work carried out in Russia, Poland and Georgia (Perepanova et al 1995; Ceyssens et al., 2009; Górski et al., 2009; Kutter et al., 2010; Abedon et al., 2011; Maura and Debarbieux, 2011; Chanishvili, 2012). Recently, some successful studies indicating the usefulness of phage therapy have been undertaken in Western countries, on animals (Wright et al., 2009; Alemayehu et al., 2012).
At the same time, even in countries where phage therapy is recognized, the available commercial phage mixtures have still not replaced the use of antibiotics. One possible reason relates to safety concerns associated with modern therapeutic phage mixtures, and in particular to procedures for the enrichment of such mixtures with new phages. Conventionally, enhancement of the lytic activity of phage preparations is achieved through the incubation of phage-resistant pathogenic bacteria with samples from a variety of natural sources of phages (soil, ponds, sewage, etc.). Thus, there arises a possibility that temperate phages, which are considered as unacceptable in therapy (because of their active role in the evolution of virulence and pathogenicity) (Fortier and Sekulovic, 2013), may become included in therapeutic products during a process of uncontrolled enrichment. Usually, the spectrum of lytic activity is considered to be an important criterion of the efficacy of a phage preparation. True efficacy, however, may depend also on the final level of era dication of bacterial cells in the microbiota at the location of the infection.
Consequently, the only possibility for developing safe phage therapy is through the use of mixtures of well-studied phages, based not only on the results of genome annotations of selected phages, but upon a comparison with other phages of the same species. As will be shown in the results of the present work, even phage genomes that are almost identical in structure and composition can differ in important respects, raising doubts as to the possibility of using phages of some particular species.
Such an approach requires an in-depth study of the candidate phages that are available. Unfortunately, there is no obvious way in which the precise functions of all the gene products of a phage can be elucidated in order to identify 'undesirable' genes. The problem can be only partly addressed by phenogenetic studies of phages involved. The use of PCR (Polymerase Chain Reaction) or metagenomic analysis cannot provide guarantees of the safety of phage mixtures produced by the method of random enrichment. However, in some cases, there is the potential to use simple procedures based on visual comparisons for rapid evaluation of various properties of phages that are relevant to their use in therapy (see Results).
The use of phage mixtures leads to the emergence and dissemination of multiple-phage-resistant (MPR) strains. To prevent this, constant adaptation of lytic activity of phage mixtures for actual bacterial strains is required. It is unclear at present whether the extent of variability of the various different phage species is sufficient to support the rapid isolation and substitution of phages with new lytic spectra. As will be shown later, not all lytic phages with a good host range are acceptable for use in therapy.
It is sometimes suggested that the set of available therapeutic phages might be enlarged by the addition of lytic variants of temperate phages, even transposable ones (Kim et al., 2012). As is well known, temperate phages can be active participants in the construction of pathogenic islands in bacterial chromosomes (Miao and Miller, 1999; Faruque et al., 2003; Tinsley et al., 2006; Winstanley et al., 2009). Genomes of transposable temperate phages that are integrated into conjugative plasmids are able to migrate across a wide range of bacterial species. The use of such specific vectors for genetic exchanges between unrelated bacterial species is capable of creating new bacterial strains with unexpected and potentially dangerous properties (Chaconas et al., 1981; Plotnikova et al., 1982 and 1983; Jenkins et al., 1985; Groisman and Casadaban, 1987; Kaplan et al., 1988). The activity of transposable phages of P. aeruginosa is a basic cause of the emergence of the highly aggressive epidemic of P. aeruginosa strains in cystic fibrosis centers worldwide (James et al., 2012). Thus, the use of nat ural lytic variants of any temperate phage is unacceptable.
Som etimes it is difficult to denote the phage as either virulent or temperate. The term "pseudolysogeny" is frequently used to refer such cases, in which the nature of the interaction of the phage with sensitive bacteria is non-obvious. There are different reasons for pseudolysogenic condition. For instance, a study of the interaction between temperate phages and bacteria in chemostat continuous culture (Ripp et al., 1998) showed that under conditions of starvation and slow growth of bacterial cells, there was an increase in the frequency of pseudolysogenic cells (cells that were infected but in which there was no intracellular development of phage), but that when nutrients were sufficient, there occurred a choice in favor either of lysogenization or of lytic phage development. This may be considered as a specific strategy of bacteriophages to survive periods of starvation and thus to prevent abortive development. This strategy can be put to advantage in the selection of bacteriophages suitable for the treatment of infections in which the pathogens form stable biofilms. Bacterial growth activity in a biofilm decreases and some phages may lose efficacy as a consequence, even though their lytic activity may be excellent on fresh bacterial lawns in Petri dishes.
The other situation arises as a result of true but unstable lys ogeny and is a consequence of the weak activity of the particular phage repressor concerned. We will show that for P. aeruginosa phages, both situations may be encountered. Hence, since the use of "natural" phages can be the source of serious problems, each new phage proposed for phage therapy should be carefully studied, not only by genome sequencing and annotation, but in a subsequent obligatory and detailed analysis of its development in different bacterial hosts. As a result, not only lytic variants of transposable phages but phages that produce pseudolysogenic conditions may be completely excluded from use in therapy. This approach therefore addresses two issues that are vitally important for phage therapy: 1) What is the level of knowledge of a phage that is necessary and sufficient to ensure its safe use in therapy? 2) Of the phages that are already known in existing collections, which of them can be used without possible complications?
-
Pseudomonas aeruginosa strain PAO1, received from Prof. B. Holloway (Monash University, Melbourne, Australia), was used routinely as the host for most of the phages mentioned in the experiments described below. P. aeruginosa strain Pu21 (pMG53) (from Prof. R. Miller, University of Tennessee, Knoxville, USA) was used as the host for phages TL and CHU (see below). P. aeruginosa strain 8-20s (from Prof. C. Pourcel, Université Paris-Sud, Orsay, France) selectively supports the growth of CHU (i.e. not the growth of TL). Clinical isolates were received from a number of clinics in Moscow (Russian Federation). The mucoid strain P. aeruginosa Pse163 is from Professor M. Vaneechoutte, University of Ghent, Belgium.
-
P. aeruginosa bacteriophages of species phiKZ (NC_004629), EL (NC_007653), RU (not sequenced), TL (NC_023583.1), CHU (sequencing in process) and phiKMV (NC_005045.1), and their variants, had been isolated and studied previously in our laboratory (Lavigne et al., 2003; Krylov et al., 2010; Pleteneva et al., 2011). All other bacteriophages used in this work were from our own laboratory collection, the Lindberg phage-typing set of P. aeruginosa phages (Lindberg and Latta, 1974) and the B. Holloway collection (Holloway et al., 1960).
-
Standard nutrient media and conditions were used for the culture of bacteria and bacteriophages (Sambrook et al, 1989).
-
Bioinformatic analysis of DNA sequences was carried out using the Basic Local Alignment Search Tool (BLAST) from NCBI (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi).
Bacterial strains
Bacteriophages
Media and conditions for incubation
Bioinformatic analysis
-
There is an accepted general protocol for the characterization of a newly isolated phage, viz.: selection of the phage from the external source / DNA isolation / genome sequencing / genome annotation / and a recommendation that the phage is suitable for use in therapy. The biological characterization of phages that are deemed suitable for use in therapy is usually limited to an estimation of their host range, and their designation as temperate or virulent. Some further features can be identified from an annotation of the genome. However, only a direct comparative study of a new phage in relation to other phages including visual observation of phages interactions on the surface of infected bacterial biofilm, can provide a reliable indication regarding its safety for therapeutic application. This is now illustrated below, using some new findings.
Until June 2014, the NCBI database contained information on 66 P. aeruginosa phages in the Caudata group. Most of these phages (47 out of 66) are considered as virulent and can be classified into a small number of species. It is evident that new species will be found, but it is also clear that the number of such new species will not be excessive. The presently known various species of P. aeruginosa virulent phages are presented in Table 1. All in all, there are 11 species of supposedly virulent phages for which the genomes have been annotated (genome of Lin68 has not been sequenced yet).

Table 1. Summary of species of virulent bacteriophages active against P. aeruginosa
The species that are evident candidates for use in therapy are the first five in Table 1, which are also the most frequently occurring: the PB1-, phiKMV-, PaP1-, KPP10-and PaP3/LUZ24-like phages. The frequency of their isolation may reflect their wide lytic spectra. We detected these species of phages in three different commercially available phage mixtures (produced by Microgen in Perm, Ufa and Nizhnii Novgorod – all in the Russian Federation). We have noted that some of the phages from these commercial mixtures exhibit a specific phenotype (opales cence) that may be related to that of phage phiKZ and other giant phages. As it transpires, phages of species phiKZ are permanent components of commercial mixtures, although EL-like and Lin68-like phages have not been yet found.
It is noteworthy, that in general, the sizes of phage DNAs of every species have their specific confined range. The largest volume may reflect the maximum capacity of the capsid, and the smallest may correspond to the size of the DNA molecule that is required in order to contain all of the important phage genes for that species, although there may be specific exceptions (Sokolova et al., 2014). The s ize difference between the DNAs of phages within the same species perhaps suggests the presence of excessive nucleotide sequences that are not an essential component of the phage genome. This might imply that phages which carrying minimum number of ORFs open preferable for use in therapy.
-
phiKMV-like phages are present in all commercial preparations. DNA restriction analysis for several phages reveals an appreciable degree of similarity to phiKMV (Burkal'tseva et al., 2006). It would appear that their presence in commercial phage mixtures reflects their high growth rate and their wide range of lytic activity. According to data (Ceyssens et al., 2011), the duration of their latent period is 21-28 min, and their spectrum of lytic activity ranges from 5% to 58% when assessed against more than 100 randomly selected clinical isolates of P. aeruginosa. In both size and structure, the genomes of phages of this species are very conservative and DNA homology is at the level of 83–97%. It is believed that the genomes of phiKMV-like phages have emerged to a substantial extent as a consequence of vertical evolution. In some cases, the differences between genomes may be so minimal as to be detectable only by a detailed comparison of the nucleotide sequences (Kulakov et al., 2009).
We have observed that some phiKMV-like phages, e.g. phiNFS (Figure 1) present in commercial preparations, when plated on a lawn of P. aeruginosa strain PAO1 (which is considered to be a Standard host for P. aerugunosa phages), exhibit instability, with segregation of secondary mutants occurring as a result of adaptation to the new host. The secondary mutants are stable and show plaque morphology similar to that of phiKMV.
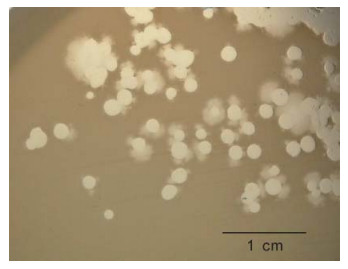
Figure 1. Phage phiNFS isolated from a commercial mixture and plated on a lawn of P. aeruginosa strain PAO1, showing the presence of plaques of unusual morphology. Repeated subcultures confirmed phage instability under these conditions. The scale bar is 1 cm.
Some newly revealed properties of phiKMV-like phages, described below, permit a deeper understanding of the interrelations of phiKMV-like phages with P. aeruginosa and alter the perception of these phages as promising candidates for use in phage therapy. Following a single day of incubation on lawns in Petri dishes, phiKMV-like phages produce large clear plaques with narrow halos. With continued incubation, however, an increasing number of bacterial colonies can be seen. In a plaque of phage phiKMV after three to four days of incubation, range of bacterial colonies can be seen, varying in appearance (Figure 2A). Two types of colonies isolated from such plaques produced phage after several repeated cycles of re-plating (Figure 2B). We consider this phenomenon to reflect a transition of the bacterial cells into a pseudolysogenic state. It is may be due to an infection of some survivors within the plaques that are in a specific physiological state.
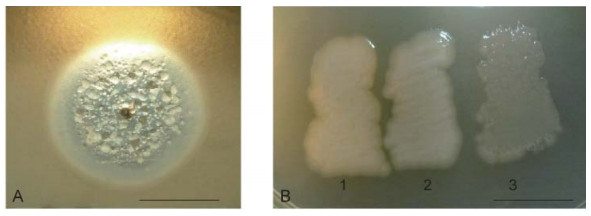
Figure 2. Pseudolysogeny of P. aeruginosa PAO1 cells after phiKMV infection. The growth of phiKMV (four days of incubation) on a PAO1 lawn is shown. The different types of resistant and sensitive colonies in the zone of lysis (A). Some of the isolated clones are phiKMV-resistant and do not produce phage (B-1); they do not affect the growth of most other phage species, including phage PB1. This means that the resistance of such mutants is not associated with loss of the surface lipopolysaccharide (LPS) (Jarrell and Kropinski, 1977). Other clones with different colony phenotypes produce phage after several re-plates (B-2 and B-3). The scale bar is 1 cm
Within a few days, the sizes of phiKMV plaques increase. Thus, unlike other phage species, phiKMV is capable of ov ercoming for a period of time the conditions occurring in an a ging biofilm, which prevent the growth of phages of other species (Figure 3). Growth of phages of several other phage species produces halos of different sizes and appearances. The halo of phiKMV phage is narrow in comparison with the halos of other phages. Halos that are especially large and mucous are formed around phiKZ-like phages (phiKZ, Lin68 and phi10/2). The formation of halos by P. aeruginosa phages is caused by the activity of depolymerases that act on polysaccharides. These enzymes, produced by a range of phage species, are structural components of the tail (Castillo and Bartell, 1974; 1976) and participate in the adsorption of phage particles to lipopolysaccharides (LPS) in the bacterial cell wall.

Figure 3. The growth of phage KMV and phages of other species from our collection after four days of incubation on a P. aeruginosa PAO1 lawn. The different interactions of the halo-producing enzymes of the various phages can be seen. They confirm the differences in their specificities, which is important in selecting the composition of phage mixtures to achieve a maximal biofilm-disrupting effect. The scale bar is 1 cm.
It is useful to compare the differences in growth patterns of phages belonging to different species in areas of confluent growth, so as to estimate the potential compatibility of the phages concerned, with a view to designing phage mixtures that can exert an optimal lytic effect in biofilm without the drawback of mutual growth inhibition. Figure 4 demonstrates the incompatibility of some phages.
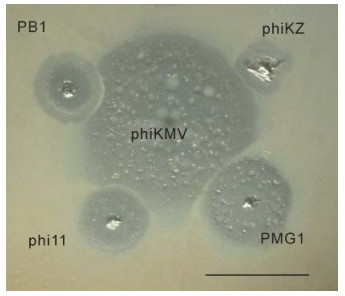
Figure 4. Mutual incompatibilities of phages, on examples shown between phiKMV (in center) and PB1 (upper left), phiKZ (upper right), phi11 (lower left), PMG1 (lower right). The scale bar is 1 cm.
Currently, it is thought that phage enzymes that contribute to halo formation during the growth of phages on lawns of non-mucoid P. aeruginosa strains can be assumed to be enzymes that are specific for the disruption of LPS in the external part of the bacterial cell wall. On the other hand, Hanlon and co-authors (Hanlon et al., 2001) have shown that although purified temperate bacteriophage F116 is capable of migrating through P. aeruginosa biofilms and that this may be facilitated by a reduction in alginate viscosity, the source of the enzyme may in fact be the bacterial host itself.
The authors of one study (Glonti et al., 2010) have suggested that a member of the phiKMV-like phages, PT6, produces an enzyme that depolymerizes the alginic acid capsule. The genome of PT6 is as yet unse quenced and unannotated, but in the annotated genomes of other phiKMV-like phages there are no genes that encode an alginate lyase. It will of interest to continue this line of enquiry, however. The possibility cannot be excluded that phage PT6 infection stimulates the production of a new bacterial alginase that is capable of destroying acetylated alginate. Further studies are clearly necessary to clarify this issue, given that the formation of acetylated alginate is the main precondition for the production of a stable biofilm of P. aeruginosa. In order to prove the hypothesis that a phage produces an alginate lyase, it is necessary to use bacterial strains that generate an excess of alginate. In experiments with strain P. aeruginosa Pse163, which produces such an excess of alginate, we could not detect evidence of anti-alginate activity in any of several phages that were tested. Thus, following infection of the stable alginate-producing strain Pse163 with phiKMV, there were no visible halos around the spot of phage after three days of incubation (Figure 5) (see also in Krylov and Shaburova, 2012).
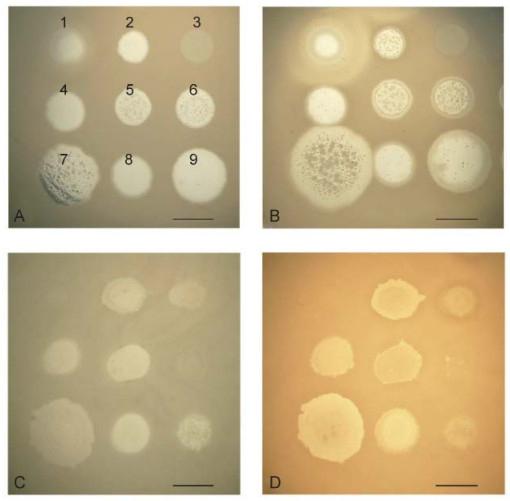
Figure 5. Comparison of the growth of several phages after overnight incubation and after three days of incubation on lawns of P. aeruginosa strains PAO1 (A, B) and PSE163 (C, D). The disposition of the phages on the bacterial lawns is given in the first panel: 1-SL2; 2-Lin68; 3-EL; 4-φC17; 5-D3112; 6-B3; 7-phiKMV; 8-E79; 9-phiMK. The spot of phiKMV is greatly increased on lawn PAO1 (A, B) with increased incubation period. The sizes of the spots for the other phages remain unchanged. On the PSE163 lawn (C, D), none of the spots for the phages, including KMV, have increased in size. Thus, phage phiKMV, an common with the other phages, is incapable of digesting the alginate of the permanently alginate-producing strain, PSE163. The scale bar is 1 cm.
We have two contradictory views on the value of phiKMV for phage therapy. Although the ability of phage phiKMV to lyse bacteria in aging biofilms can be considered as a highly useful feature, it cannot be excluded that there is an intrinsic relationship with pseudolysogeny (as it is seen on Figure 2) and, as a result, with the survival of infected bacteria in the microbiota of the infection site. Perhaps this special feature of phiKMV-like phages has been the cause of the failure to use them for the eradication of P. aeruginosa in a mouse model of cystic fibrosis (Henry et al., 2013).
Seemingly, should it prove impossible to select mutants of phiKMV-like phages without pseudolysogenic effects, it will be advisable to abandon the use of phiKMV-like phages as therapeutic agents.
-
PB1-like phages are a usual component of phage therapeutic commercial mixtures. This species is considered to be one of the most promising for application in phage therapy. One of its major advantages is the ability to generate mutants with extended lytic activity (Pleteneva et al., 2008 and 2009; Ceyssens et al., 2009), an absence of pseudolysogenic effects (complete lysis of infected bacteria), and low frequencies of phage-resistant bacterial mutants. Phages of this species are adsorbed onto LPS and produce a halo around the plaque as a result of the activities of LPS-destroying enzymes that are synthesized by bacteria in the course of phage infection.
-
Phages of species PaP3/LUZ24-like phages are permanent components of commercial mixtures; but in some of them, we found unusual properties. Their possible influence on the behavior of the phage during the process of phage therapy is not clear. Further comparative studies are required before phages of this species can be introduced into phage therapy as components of mixtures that are well-studied and safe for extended use. The first phage of this species to be described, PaP3, was isolated as a temperate phage (Tan et al., 2007). However, no repressor gene has been found in its genome. The other phage of this species, LUZ24, shows a high degree of relatedness to PaP3 and also does not encoding repressor protein. A remarkable property of LUZ24 is the presence of an intron in its genome (Ceyssens et al., 2008). The isolation of phages of related species, infecting other pseudomonads, P. putida and P. fluorescens (Glukhov et al., 2012; Eller et al., 2014), suggests a possibility for the migration of these phages between different soil pseudomonades.
Two new phages, TL and CHU, were isolated in our laboratory from natural sources. According to the results of genome sequencing, TL is closely related to other phages of the species. But, unlike other related phages, it encodes a transposase. Phage CHU shows a positive PCR response with TL primers, and produces identical fragments after DNA digestion by endonucleases. A feature common to TL and CHU is weak growth on lawns of the Standard P. aeruginosa strain PAO1. However, they grow well and selectively on lawns of some clinical isolates of P. aeruginosa (P. aeruginosa strain 8-20) or of mutants of PAO1 (P. aeruginosa strain PAO-ELR2), showing a high level of instability (Figure 6); however, the role of transposase in phage TL instability is not evident. Introduction of the plasmid pMG53 (IncP2) into P. aeruginosa strain PAO1 restores the growth of both phages, with simultaneous loss of instability. From the point of view of their use in phage therapy, this is a desirable feature because IncP2 group plasmids frequently inhibit the growth of various phage species. In addition, in the regions of the halos formed during the growth of various other phages on lawns of P. aeruginosa strain PAO1, the lytic activity of phage TL increases significantly, thereby allowing the identification of the halo-forming phages (Pleteneva et al., 2011). CHU, on the hand, does not exhibit this behavior (Figure 7).

Figure 6. Genetic instability of plaque morphology in TL (A) and CHU (B) phages, as revealed on P. aeruginosa strain PAO-ELR2 (for TL) and clinical strain P. aeruginosa 8-20 (for CHU). The scale bar is 1 cm.
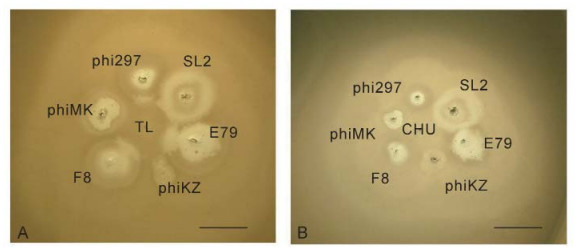
Figure 7. The growth of TL (A) and CHU (B) in the halos of some halo-forming phages on a lawn of P. aeruginosa strain PAO1. The phage TL acts as a "developer" of halos of phages phi297, E79 and F8 when grow in close proximity to their plaques. This is related to better growth in cells in the halo (Pleteneva et al., 2011). The nature of halos, produced by phages phiMK, SL2 and phiKZ is different and they do not interact with TL. In contrast, the closely related phage CHU has no such activity. The scale bar is 1 cm.
It is unclear why phage PaP3, having no repressor in its genome, has been isolated from the bacterial strain as a temperate phage. It is possible that in this case, the bacteria were in a pseudolysogenic state. Thus, only after further studies of the cause and the consequences of the growth instability of these phages will it be possible to assess their potential utility.
-
The group of phiKZ-like phages active on now includes three species, phiKZ, EL and Lin68, and they attract the attention of investigators because of the unusual structure of their phage particles and some specific features in their infection of bacterial cells (Krylov and Zhazykov, 1978 and 1984; Hertveldt et al., 2005; Thomas et al., 2008; Pleteneva et al., 2010; Krylov et al., 2011; Cornelissen et al., 2012; Sokolova et al., 2014). The common features for all phages in this group are: 1) identical morphology and size of the phage particle; 2) the specific packaging of DNA ("inner body" -helical coil with supercoiled DNA wound around it); 3) the absence of any enzyme recognized by genome annotation as a phage DNA polymerase; 4) the ability following infection with a high multiplicity to convert all of the bacterial cells into a pseudolysogenic condition (carrier state). Supposedly, the generality of these characteristics reflects the phylogenetic relationship of the various species within this group.
Pseudolysogeny in the case of the phiKZ-like phages displays very specific features that differentiate it from the cases of pseudolysogeny previously mentioned. Thus, in a one-step growth cycle experiment following the infection of sensitive cells with a multiplicity of infection (m.o.i.) in the range of one to five particles, these phages exhibit features of virulent phages – all infected cells were killed, with the liberation of a modest number of phage particles (Krylov and Zhazykov, 1978). At higher m.o.i. values, however, infection with phiKZ-like phages leads eventually to a special state in which bacterial cells continue to divide, leading to the formation of colonies that can grow for several days in Petri dishes. In consequence, these colonies produce huge amounts of phage particles (Krylov and Zhazykov, 1978 and 1984; 2004; Burkal'tseva et al., 2002; Pleteneva et al., 2009). This may be of particular significance, in view of the fact that phages of species phiKZ have been found in various commercial therapeutic blends. Phages of species EL and Lin68 are infrequent (we have isolated two phages, RU and CHE, which are closely related to EL, and a single phage LBG22, which is related to Lin68) (Burkal'tseva et al., 2002).
To date, all the phiKZ-like phages that have been sequenced do not encode "Standard" DNA-polymerases (Mesyanzhinov et al., 2002; Hertveldt et al., 2005). Nevertheless, a very unusual DNA polymerase activity of a new type has been found to be encoded in the genomes of phiKZ-like phages (Cornelissen et al., 2012). It is possible that under the conditions that arise following the high-multiplicity infection of cells, phage development is turned off for a period of time.
The precise mechanism for such an effect of m.o.i. is not yet clear, but it is evident that such phages, containing the phage genome in its wild-type state, can facilitate HGT (because pseudolysogenic cells can support the development of other phages, including temperate phages). Thus, pseudolysogeny is one of the reasons why phiKZlike phages in their wild-type state are not desirable components of phage mixtures.
Such unusual behavior could not have been predicted from the sequencing and annotation of the phage genomes. Furthermore, a study of phiKZ transcription (Ceyssens et al., 2014) has found one other unique feature of this phage: phiKZ does not require a functionally active bacterial transcriptional system. The mechanism of temporary lysis inhibition is not yet elucidated. It is of interest that in both the phiKZ and EL genomes there exist genes that encode proteins similar to the repressors of phages that are specific for unrelated bacterial species (Mesyanzhinov et al., 2002; Hertveldt et al., 2005). These may function as a repressor-type activity, blocking the lytic cycle and leading the infected cells into a pseudolysogenic state. Bacterial cells in such a state, being infected with phiKZ, are capable of movement, division and the production of cells sensitive to phage infection (Krylov et al., 2013). In the case of cells infected with phiKZ-like phages at high m.o.i., the pseudolysogenic state offers a biological advantage (to the phage) because, in the absence of a phage-coded DNA polymerase and given that these phages are independent of bacterial transcriptional activity, pseudolysogeny leads to a great increase in phage production. Some of the cells become temporarily resistant to phage infection and are able transport phage particles (phage as a "rider") (Krylov et al., 2013).
Unconditional evidence of true but unstable lysogeny is provided by the selection of mutants with properties similar to those of virulent mutants of temperate phages (Krylov et al., 2011). Such virulent variants kill cells that are in a pseudolysogenic condition. The use of such mutants in therapeutic mixtures instead of wild-type phiKZlike phages will prevent the possibility of HGT (Krylov et al., 2010; Pleteneva et al., 2010; Krylov et al., 2011). The dominance of mutant phage phiKZ in mixed infections with EL shows that even in choosing between related phages for phage-therapy application, it is necessary to take into attention the possibilities of mutual inhibition (Krylov et al., 2013).
Excellent evidence that phiKZ-like phages cannot be used in phage therapy in their wild-type state has been reported in a recent study (Henry et al., 2013) that showed that it was impossible to eradicate P. aeruginosa in a mouse-lung infection model.
Gener al description about virulent bacteriophages of P. aeruginosa
phiKMV-like phages and features of their pseudolysogenic growth in biofilms
Species of PB1-like phages
Species of PaP3/LUZ24-like phages–observation of genetic instabilities
The group of phiKZ-like phages: pseudolysogeny and selection of virulent mutants
-
The increase in the number of infections caused by multidrug-resistant pathogenic strains provides a stimulus to the development of alternative approaches, including phage therapy. However, phage therapy is not yet a generally accepted method of anti-bacterial therapy, even in those cases where it could replace antibiotics, and even in those countries where it is permitted. The reasons for this are diverse, but they include a degree of conservatism amongst physicians and a lack of essential information.
Currently, there are no generally accepted and detailed criteria and protocols for the use of phage preparations. The reason may in part be uncertainty and concern regarding the safety and reliability of phage therapy. Such fears are justified, because the procedures now used for updating the composition of phage mixtures do not preclude the inclusion of temperate phages if they extend the lytic activity range of the preparation. Obviously, to conduct a reliable study of a phage mixture obtained by random enrichment (which can be denoted as "phage products of the first generation") is not straightforward; PCR is not adequate for this. Metagenomic analysis could provide an overall assessment of the phage composition and, as a result by implication, its safety in therapeutic use (McCallin et al., 2013). However, this should be done for each batch of product made following any enrichment by the introduction of new natural phages. The positive outcomes of such a test should then increase the confidence of doctors and patients in the safety of "phage products of the first generation".
The use of complete personalization of phage therapy (meaning the isolation directly from the environment of phages active against a specific pathogen) will improve its safety and efficacy (Henry et al., 2013). However, in this case, there is a requirement to undertake a full study of individually selected phages within tight time constraints. This approach can be seen as desirable in the phage therapy of chronic pulmonary infections in patients with cystic fibrosis, given the proven role of a number of phages in increasing the pathogenicity of P. aeruginosa (Winstanley et al., 2009). However, it is unlikely that complete personalization will be undertaken, for example, in the treatment of wounds or of nosocomial infections.
Instead, for the treatment of wounds and hospital infections, there is the possibility of using partial personalized therapy. In this case, the therapeutic phages may be selected from previously studied bacteriophages in existing collections. This need not preclude the possible use (under conditions designed to prevent completely the spread of phages beyond the ward designated for phage therapy) not only of virulent phages but also of lytic variants of temperate phages, such as PMG1, the natural lytic variant of phage D3, or the natural lytic phage YMC01/ 01/P52 PAE BP, related to phi297 (Jeon et al., 2013).
We believe that the time has now come to progress to the creation of "Phage Therapy Products of the Second Generation" (PTP-2). In this case, a mixture with maximal lytic activity range will be compiled selecting only previously studied virulent phages of various species. It will be possible to add into such mixtures, comprising lytic phages, variants of pseudo-temperate phage species (of phiKZ-like species–phiKZ and EL) or of phage species PaP3/LUZ24 and phiKMV (provided that it is possible to isolate suitable variants that lack the ability to cause bacterial pseudolysogeny and that do not show manifest genetic instability).
The few simple criteria for the assessment of phages from the viewpoint of practical suitability for therapy that have been presented here (identification of phages causing the pseudolysogenic state in infected bacteria, phages exhibiting signs of mutual exclusion/incompatibility and selection of phages for optimal disruption of capsular polysaccharides) may simplify the process of compounding effective lytic therapeutic preparations, avoiding the need in some cases to check the attributes of phages in animal models. The basic and as yet unsolved problem in phage therapy is the lack of knowledge of the functions of most of the gene products encoded by the phage genome. There is always the possibility that the genome of a phage of a species that is recognized as suitable for use in phage therapy will contain a gene encoding an undesirable product (such as the transposase of phage TL in species PaP3/LUZ24). Using comparative genetic studies of groups of related bacteriophages, it is possible to identify such "problem" genes, to estimate their importance for the viability of the phage and to inactivate them by site-directed mutagenesis. The transition to the "Third Generation of Phage Therapy" will depend on the development of 'recombineering', genetic modification techniques based on the use of such recombinases as lambda phage enzyme Red, capable of performing recombination using very short regions of homology (Murray 2006; Fehér et al., 2012; Marinelli et al., 2012; Schmelcher et al., 2011; Thomason et al., 2009). The adaptation of recombineering procedures for use with DNA of P. aeruginosa phages will permit their genomes to be tailored, permitting the use of modified phages in safe and totally controlled therapies – but that is another story.
Once again, it is necessary to emphasize the number one rule in any type of phage therapy: to prevent the accidental d issemination of bacteriophages beyond their designated place of use-called as "Phage Hospital Ward" (PHW) – to the parts of the hospital (Krylov et al., 2014).
-
The authors express their gratitude for collaboration in phage studies to Prof. Leonid Kulakov (UK), and to Prof. Cristine Pourcel (France), and Prof Mario Vaneechoutte (Belgium) for bacterial strains used in the study.
-
All the authors declare that they have no competing interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
VK designed the experiments. OS, EP, AK, MB and OP carried out the experiments. OS, SK and MB analyzed the data. VK and OS wrote the paper. All authors have read and approved the final manuscript.













 DownLoad:
DownLoad: