HTML
-
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a novel coronavirus that causes the outbreak of coronavirus disease 2019 (COVID-19) (Li et al. 2020a). Viral nucleic acid testing is the standard method for the laboratory diagnosis of COVID-19 (Wu et al. 2020a; Zhu et al. 2020). Currently, a variety of qPCR-based detection kits are used for laboratory-based detection and confirmation of SARS-CoV-2 infection (Corman et al. 2020; Hussein et al. 2020; Ruhan et al. 2020; Veyer et al. 2020). Conventional qPCR involves virus inactivation, nucleic acid extraction, and qPCR amplification procedures. Therefore, the process is complicated, which usually takes longer than two hours, and requires biosafety laboratories and professional staff. Thus, qPCR is not suitable for use in field or medical units. To reduce the operation steps, automatic integrated qPCR detection systems that combine nucleic acid extraction and qPCR amplification in a sealed cartridge were developed to detect viruses in clinical samples (Li et al. 2020b). However, the detection time is still longer than one hour. Therefore, rapid nucleic acid detection systems are needed to further improve the detection efficiency.
Recombinant polymerase amplification (RPA) is a novel isothermal amplification technique that can amplify a target gene within 15–20 min at room temperatures (25–42 ℃). Compared with qPCR, the amplification reaction is faster and the equipment is both compact and cheap, making it more suitable than qPCR for field application (Piepenburg et al. 2006; Wang et al. 2017; Jia et al. 2019). Behrmann and Wu developed RPA-based assays to detect SARS-CoV-2 nucleic acid (Behrmann et al. 2020; Wu et al. 2020b). However, in their system, samples still need to be inactivated and the nucleic acids need to be extracted in a BSL-2 biosafety laboratory prior to RPA amplification. Therefore, no RPA detection system is available for the rapid direct detection of raw sample.
In this study, we developed a rapid integrated-RPA (I-RPA) system to detect nucleic acids from clinical samples. The method comprises an integrated rapid sample treatment and nucleic acid isothermal amplification process within a single sealed cartridge. No additional nucleic acid extraction processes are required. The time from adding the raw sample to the cartridge to obtaining the result is 30 minutes.
The I-RPA system comprises a cartridge and an automatic nucleic acid detection device. The cartridge consists of three areas (placing the treatment buffer, catalyst MgAc and RPA reaction mixture respectively), and the adjacent areas are separated by a plunger seal. The I-RPA procedure flow chart is shown in Fig. 1A. and the detailed method is shown in Supplementary Materials.
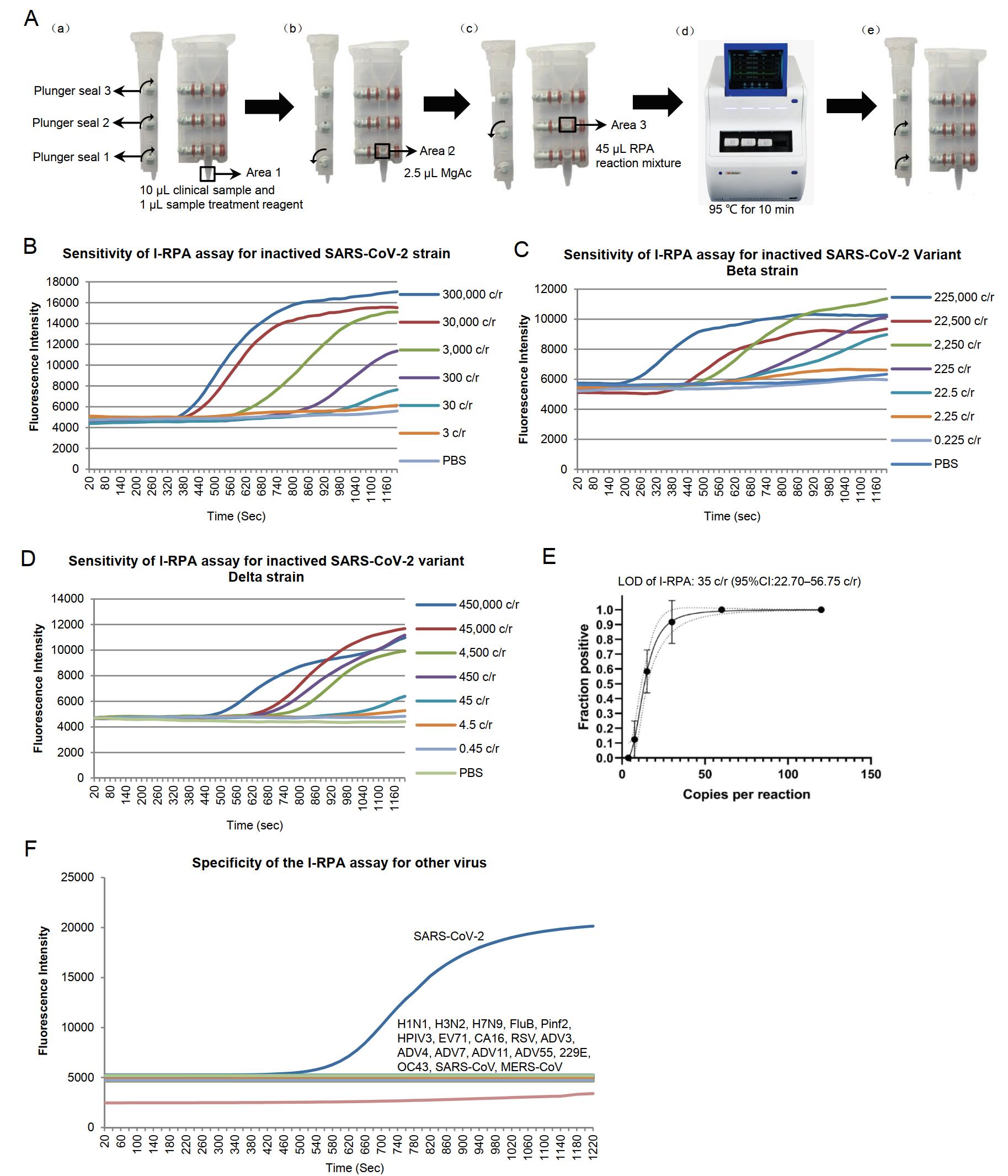
Figure 1. A Flow chart of the I-RPA system. The cartridge consists of area 1, area 2, and area 3. Adjacent areas are separated by plunger seals. A-a: The plunger seals in the cartridge are opened and 10 μmL of each clinical sample is mixed with 1 μmL of sample treatment reagent (TCEP and EDTA mixture) and added to area 1. A-b: Plunger seal 1 is closed, and 2.5 μmLof MgAc is placed in area 2. A-c: Plunger seal 2 is closed, and a total of 45 μmL RPA reaction mixture comprising 2.1 μmL of forward primer (10 μmmol/L), 2.1 μmL of reverse primer (10 μmmol/L), 0.6 μmL of probe (10 μmmol/L) and 40.2 μmL of rehydration buffer A (Zhongce Ltd., Hangzhou, China) is added to area 3. A-d: Plunger seal 3 is closed. The lid of cartridge is closed and the cartridge is placed into the I-RPA device (LifeReady1000, Hangzhou Lifereal Biotechnology, Hangzhou, China). The cartridge is heated at 95 ℃ for 10 min. A-e: The cartridge is removed from the machine, plunger seals 1 and 2 are opened. Swing the cartridge to let the liquid in areas 2 and 3 mixed in area 1, and then the cartridge is put back into the I-RPA device and incubated at 42 ℃ for 20 min. During the I-RPA assay, real-time fluorescence curve were shown in the screen of the I-RPA device, and the results (positive or negative) was analyzed automatically and shown in the screen after the I-RPA assay finished. B-D The sensitivity analysis of the I-RPA assay for SARS-CoV-2 BetaCoV/Beijing/IME-BJ05/2020 (B), SARS-CoV-2 Beta variant strain CSTR.16698.06.NPRC 2.062100001 (C) and Delta variant strain CSTR.16698.06.NPRC 6.CCPM-B-V-049-2105-6 (D). The viral RNA were extracted from culture supernatant and ten-fold diluted. E Limit of detection (LOD) of the I-RPA assay for SARS-CoV-2. The x-axis shows the amount of input virus per reaction. The y-axis shows the positive percentage for all parallel reactions performed. The LOD is shown in the panel headings. The inner line is a probit curve (dose-response rule). The outer dotted/dashed lines are the 95% confidence intervals. F Specificity of the I-RPA assay for inactived respiratory tract-associated virus cultures. For the I-RPA assay, the Real-time fluorescence signals were documented over time. The fluorescence signal threshold was set as three standard deviations above the background signal determined within minute 1; the slope (mV/min) was calculated by AIGS software. Signals were interpreted using combined threshold and signal slope analysis. A positive reading was set as fluorescence signal higher than the fluorescence signal threshold and the slope greater than 15 mV/min within 20 min.
For the design of primers and probes, ten genome sequences including SARS-CoV-2 strains and variant strains were downloaded from the GISAID website (https://www.gisaid.org). These sequences were aligned by using MegaAlign software and a consensus sequence of the SARS-CoV-2 spike gene was obtained (Supplementary Fig. S1). Specific primers and probe targeting this sequence were designed and then synthesized by Sango (Shanghai, China). The length of the RPA primers and probe was set to 30-35 nucleotides (nt) and 50 nt, respectively. Nine combinations of candidate primers (three forward primers and three reverse primers) were prepared to screen the highly sensitive primer set (Supplemental Table S1).
RNA extracted from inactivated SARS-CoV-2 BetaCoV/Beijing/IME-BJ05/2020 strain, SARS-CoV-2 Variant Beta CSTR.16698.06.NPRC 2.062100001 strain and SARS-CoV-2 variant Delta CSTR.16698.06.NPRC 6.CCPM-B-V-049-2105-6 strain were used as templates. The viral RNA in those cultures was quantified by qRT-PCR using a specific in vitro-transcribed RNA quantification standard (Xu et al. 2020). The concentration of RNA extracted from BetaCoV/Beijing/IME-BJ05/2020 culture supernatant was 1.2×106 copies/μL, the concentration of RNA extracted from Beta CSTR.16698.06.NPRC 2.062100001 strain was 9×105 copies/μL, and the concentration of RNA extracted from Delta CSTR.16698.06.NPRC 6.CCPM-B-V-049-2105-6 strain was 1.8×106 copies/μL. Since the RNAs were extracted from 200 μL of viral culture supernatant and eluted with 50μL elute buffer, the gene concentrations of the three viral culture supernatants could be calculated as 3×108 copies/mL, 2.25×108 copies/mL and 4.5×108 copies/mL.
The RNA extract from BetaCoV/Beijing/IME-BJ05/2020 culture supernatant was serially diluted 10-fold (with the concentration of 1–1000 copies/μL) and used as template to select the most sensitive primer set for the I-RPA assay. Of the nine primer sets tested, six (F2R1, F2R2, F2R3, F3R1, F3R2, and F3R3) yielded positive results (Supplementary Fig. S2). Primer set F3R2 showed the highest sensitivity with the probe, with a limit of detection (LOD) about 20 copies/reaction. Consequently, primer set F3R2 was selected for the I-RPA assay.
To test the sensitivity of the I-RPA assay, viral culture supernatants were diluted 10-fold (from 3×108 copies/mL to 3 copies/mL for the BetaCoV/Beijing/IME-BJ05/2020 strain, 2.25×108 copies/mL to 2.25 copies/mL for the variant Beta CSTR.16698.06.NPRC 2.062100001 strain, and 4.5×108 copies/mL to 4.5 copies/mL for the variant Delta CSTR.16698.06.NPRC 6.CCPM-B-V-049-2105-6 strain). A total of seven different concentrations for each strain were used as templates for I-RPA assay. All samples were tested with adding 10 μL aliquots of templates in each reaction. The results revealed that I-RPA generated positive results in test samples containing > 3000 copies/mL of BetaCoV/Beijing/IME-BJ05/2020, 2250 copies/mL of variant Beta strain, 4500 copies/mL of variant Delta strain, suggesting the sensitivity of this I-RPA for the variant SARS-CoV-2 strains were similar with the original strain, and the LOD is about 30 copies per reaction (Fig. 1B–1D).
Next, to determine the LOD of this I-RPA for SARS-CoV-2, the viral culture supernatant of the BetaCoV/Beijing/IME-BJ05/2020 strain was used as representative template and diluted 2-fold (3×103–187.5 copies/mL) to yield five different concentrations around the detection end point determined in preliminary dilution experiments for I-RPA assay, three individual replicated reactions and eight replicates for each sample at each experiment were done for the LOD assay. Probit regression analysis was performed to calculate the LOD by interpolation. Based on the Probit regression average, the LOD in the SARS-CoV-2 culture was calculated as 35 copies/reaction, with a 95% confidence interval of 22.78–56.75 copies per reaction (Fig. 1E). This level of sensitivity is similar to that of the standard RPA, but less than that of qRT-PCR, which has a sensitivity of 10 copies of SARS-CoV-2 RNA/reaction (Li et al. 2020b).
The specificity of the I-RPA assay was determined in two duplicates using a representative panel of inactivated respiratory tract-associated viruses, including adenovirus types 3, 4, 7, 11, 14, and 55 (ADV3/Xinjiang2019-1415-9 strain, ADV4/Shenyang-1815-4 strain, ADV7/Shenyang-1706-20 stain, ADV11/Shenyang2018-1817-2 strain, ADV14/Xinjiang2019-1602-1 strain, ADV55/Shenyang2018-1815-2 strain); influenza A viruses H1N1, H3N2, and H7N9 (A/California/7/2009(H1N1) strain, A/Wisconsin/67/2005(H3N2) strain, A/Anhui/01/2013(H7N9) strain); influenza B virus (FluB/Xinjiang2018-1801-03/YAM strain); parainfluenza viruses 2, and 3 (Pinf2/Xinjiang2018-1607-2 stain and HPIV-3 LZ1728 strain); Enteroviruses EV 71 and CA16 (AH/08/06(EV 71) strain and CA16-G20 strain); respiratory syncytial virus (RSV 1373 strain); coronaviruses 229E and OC43 (CoV229E/Xinjiang2018-1750-2 strain and CoV43/Xinjiang2018-1749-3 strain). All of the virus trains were stored in the Academy of Military Medical Science Institute of Microbiology and Epidemiology. Vitro synthesized spike gene fragment of SARS-CoV (GenBank: AH013657.2) and MERS-CoV (GenBank: MG923480.1) were also used as templates. No consistently positive signals were observed in any of these samples (Fig. 1F).
To assess the performance of the I-RPA assay for detecting clinical samples, 38 pharyngeal swab samples and 4 sputum samples from 42 different patients with COVID-19 (mainly from the Centers for Disease Control Prevention of Hubei Province) were tested. And 53 SARS-CoV-2-negative samples from Centers for Disease Control Prevention of Beijing were used as control. All the samples were inactivated and then applied to the I-RPA assay. Meanwhile, RNA extracts from these samples were also analysed in the qRT-PCR assay (Supplementary Materials). All of the tests conducted in two separate experiments. All the 53 SARS-CoV-2-negative samples were negative in both I-RPA and qRT-PCR assay. Among the 42 samples from infected patients, all were SARS-CoV-2 positive in the qRT-PCR assay. However, three of the PCR-positive samples were negative in the I-RPA assay, with the average CT values of 36.84, 37.11 and 38.69 (Table 1, Supplementary Table S2). The result demonstrated that the specificity of I-RPA was consistent with real time PCR, but the sensitivity of I-RPA was lower than that of qPCR. This might be mainly because the efficiency of isothermal amplification was not as high as that of PCR, and the impurities in clinical samples might also interfere with the efficiency of amplification.
qRT-PCR I-RPA Total Positive Negative Positive 39 3 42 Negative 0 53 53 Total 39 56 95 The results were analyzed by Kappa consistency test. Kappa = 0.936. Table 1. Detected results of clinical samples by I-RPA and qRT-PCR assay.
In summary, we explored an I-RPA method, which did not require RNA extraction. The RPA mixture, the catalyst (MgAc), and sample treatment buffer mixture were added to three different areas of a sealed cartridge, which might prevent the contamination. The LOD of the I-RPA assay was about 35 copies/reaction. Although the sensitivity of I-RPA assay is not as high as qRT-PCR and the false-negative result might occur when testing weakly positive samples (CT values > 35), it is faster than qRT-PCR (30 minutes versus 2 hour) and only requires portable equipment, which makes it very suitable for rapid screening and detection of suspicious samples in primary medical institutions. The I-RPA system can also be expanded to enable rapid screening and detection of other pathogens.
-
This work was supported by the National Major Science Program Foundation (No. JK2020NC017 & 2018ZX10711001-003) and the Scientific Research Projects of the General Administration of Customs (No.2019HK042).
-
The authors declare that they have no conflict of interest.
-
The informed consent have been obtained from all participants and the studies have been approved by the Ethics Committee of the Academy of Military Medical Science.
Conflict of Interest
Animal and Human Rights Statement
-
The online version of this article contains supplementary material, which is available to authorized users.
-
The qRT-PCR reaction contained 5 μL RNA, 5 μL of 4× TaqMan Fast Virus 1-step mix (Applied Biosystems, Vilnius, Lithuania), 1 μL of forward primer (10 μmol/L), 1 μL of reverse primer (10 μmol/L), 0.5 μL of probe (10 μmol/L), The sequences of the primers and probe was were as follows: Forward: 5'-TCCTGGTGATTCTTCTTCAGGT-3', Reverse: 5'-TCTGAGAGAGGGTCAAGTGC-3', Probe: 5'FAM-AGCTGCAGCACCAGCTGTCCA-BHQ1, and 7.5 μL of sterile deionized water; final volume was 20 μl. Reactions were performed in a LightCycler 480 Real Time PCR instrument (Roche Diagnostics, Mannheim, Germany). Amplification conditions were as follows: reverse transcription at 50 ℃ for 5 min; pre-denaturation at 95 ℃ for 10 sec; and 40 cycles of PCR amplification consisting of denaturation at 95 ℃ for 5 sec, annealing at 60 ℃ for 30 sec, and fluorescence measurement.
-
The I-RPA system comprised a cartridge and an automatic nucleic acid detection device. The cartridge (Fig. 1A) consists of area 1, area 2, and area 3, adjacent areas are separated by a plunger seal.
Sample treatment buffer consisted of TECP (0.1 μmol/L) and EDTA (0.001 mol/L). RPA reaction mixture was prepared by adding 40.2 mL of buffer A (Zhongce Inc., Hangzhou, China), 10 mL of water, 2.1 mL of forward primer (10 μmol/L), 2.1 mL of reverse primer (10 μmol/L), and 0.6 mL of the probe (10 μmol/L) to the dry enzyme tube, followed by mixing. For the I-RPA assay, the plunger seals in the cartridge were turned to the open position, and 10 μL of each clinical sample was mixed with 1 μL of sample treatment buffer (Fig. 1A-a). The mixture was added to area 1 of the cartridge, after which plunger seal 1 was closed (Fig. 1A-b). Next, 2.5 μL of MgAc was placed in area 2 and plunger seal 2 was closed (Fig. 1A-c). Finally, the RPA reaction mixture was added to area 3 and plunger seal 3 was closed. The lid of the cartridge was closed tightly and the cartridge was placed in the I-RPA device (LifeReady1000, Hangzhou Lifereal Biotechnology, Hangzhou, China) for amplification (Fig. 1A-d). The cartridge was heated at 95 ℃ for 10 min and then removed from the I-RPA device. Next, plunger seals 1 and 2 were opened and the cartridge swung so that the liquid in areas 2 and 3 flowed into area 1 (Fig. 1A-e). Finally, the cartridge was placed back in the I-RPA device and incubated at 42 ℃ for 20 min. Real-time fluorescence signals were detected and analyzed over time.
Primer name Sequence (5'to 3')a Position (size, bp)b Cov-RPA-F2 ACATAGAAGTTATTTGACTCCTGGTGATTC 22, 269–22, 298 (30) Cov-RPA-F3 TAGAAGTTATTTGACTCCTGGTGATTCTTC 22, 272–22, 301 (30) Cov-RPA-F1 TATTTGACTCCTGGTGATTCTTCTTCAGGT 22, 279–22, 308 (30) Cov-RPA-P AAAGTCCTAGGTTGAAGATAACCCACATAAFAAHCQGCAGCACCAGCTGTCCA-[3'-block] 22, 309–22, 361 (53) Cov-RPA-R1 ACACTTTGTTTCTGAGAGAGGGTCAAGTGC 22, 411–22, 440 (30) CoV-RPA-R2 TCAAGTGCACAGTCTACAGCATCTGTAATG 22, 389–22, 418 (30) CoV-RPA-R3 AACAATAGATTCTGTTGGTTGGACTCTAAA 22, 489–22, 518 (30) a F: FAM-dT, thymidine nucleotide carrying fluorescein; H: THF, tetrahydrofuran spacer; Q: BHQ1-dT, thymidine nucleotide carrying Black-Hole Quencher 1; 3'-block: 3'-phosphate introduced to block elongation.
b Numbers in position column are primer positions according to SARS-CoV-2, hCoV-19/Wuhan/WH01/2019|EPI_ISL_406798|2019-12-26Table Table S1. Sequences of the primers and probe used for the RPA assay
Sample Results Sample Results I-RPA Real-time Ct value (Mean ±2SD) I-RPA Real-time Ct value (Mean ±2SD) P1 P 27.31±0.92 P22 P 19.8±0.18 P2 P 26.98±0.33 P23 P 25.31±0.06 P3 P 29.72±0.72 P24 P 22.9±0.08 P4 P 29.81±0.07 P25 P 26.59±0.72 P5 P 29.2±0.43 P26 P 22.88±0.32 P6 P 29.07±0.78 P27 P 31.72±0.16 P7 P 28.53±0.22 P28 P 30.65±0.21 P8 P 27.66±0.56 P29 P 33.79±0.37 P9 P 31.71±0.20 P30 P 29.46±0.52 P10 P 32.9±0.14 P31 P 25.27±0.68 P11 P 29.92±0.77 P32 P 35.83±0.77 P12 P 29.2±0.65 P33 P 25.75±0.89 P13 P 29.07±0.32 P34 P 36.58±0.48 P14 P 31.59±0.35 P35 P 20.71±0.86 P15 P 33.48±0.12 P36 P 35.31±0.22 P16 P 33.14±0.42 P37 P 36±0.72 P17 N 36.84±0.68 P38 N 38.69±0.33 P18 N 37.11±0.32 S1 P 33.21±0.71 P19 P 25±1.14 S2 P 29.09±1.32 P20 P 27.88±0.98 S3 P 29.91±0.62 P21 P 17.31±1.08 S4 P 32.83±0.88 P: positive; N: negative; P1-P38: Pharyngeal swab samples; S1-S4:Sputum samples.
SD: standard deviationTable Table S2. Analysis of clinical samples by I-RPA and rRT-PCR

Figure S1. Partial alignments of oligonucleotide binding regions, SARS-CoV-2-related coronaviruses (n = 10). The panels show ten sequences of 2019-nCoV strains, aligned to the partial Spike protein sequences of hCoV-19/Wuhan/WH01/2019 beta coronavirus. The alignment also contains the India Delta variant (hCoV-19/India/CG-AIIMS-Raipur-F-81-24/2021, GISAID EpiFlu™ Database accession ID is EPI_ISL_2620743), the South Africa variant (hCoV-19/SouthAfrica/Tygerberg-461/2020|EPI_ISL_745186|2020-12-07) and the UK variant (hCoV-19/England/LOND-1267020/2020 |EPI_ISL_741243| 2020-12-11). Dots represent identical nucleotides compared with the hCoV-19/Wuhan/WH01/2019 beta coronavirus sequence. N: deleted nucleotide. Blue arrows: sequences of the primers and probe used for the RPA assay.
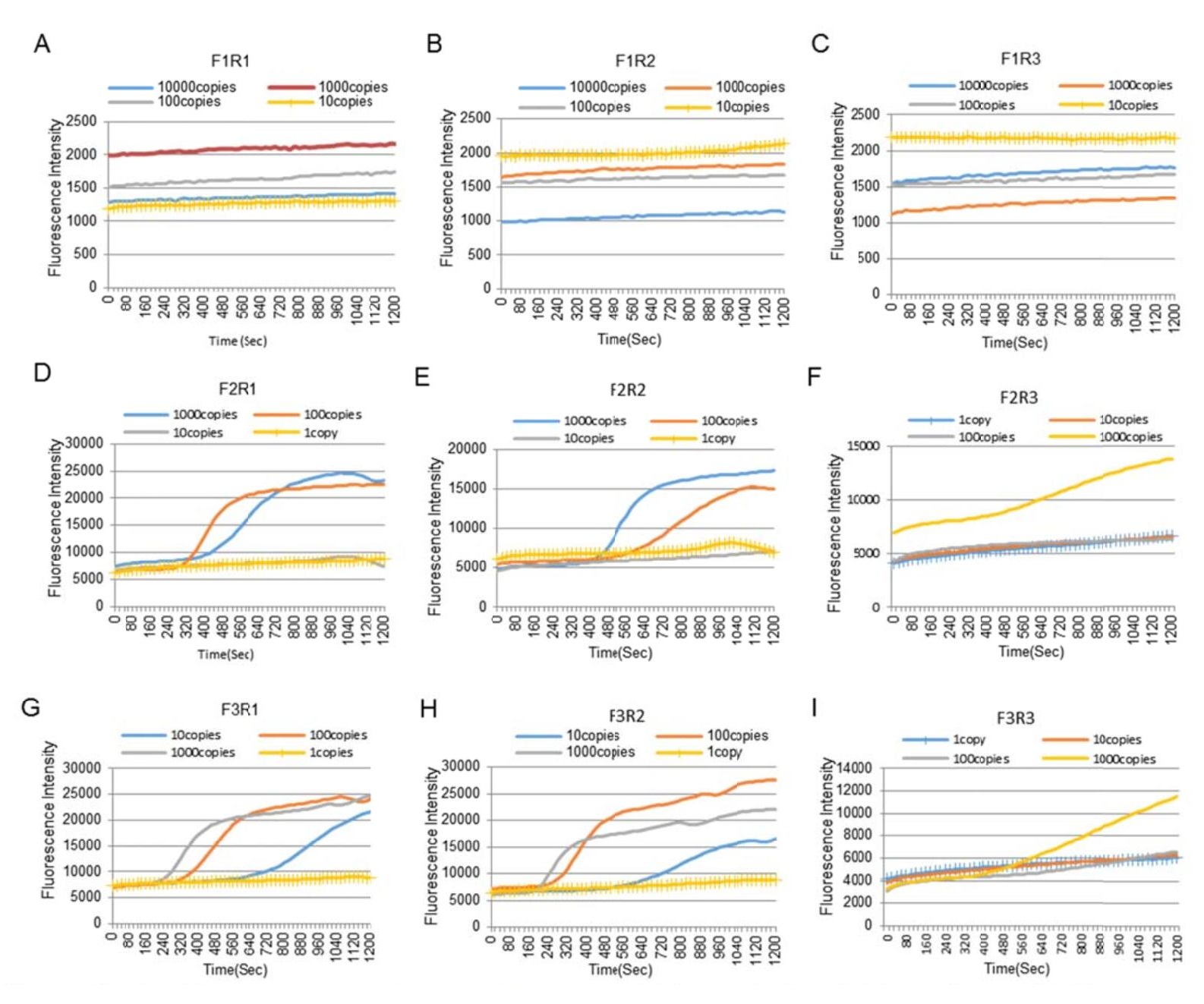
Figure S2. Sensitivity comparison of nine primer sets. (A) The sensitivity of F1R1 primers; (B) The sensitivity of F1R2 primers; (C) The sensitivity of F1R3 primers; (D) The sensitivity of F2R1 primers; (E) The sensitivity of F2R2 primers; (F) The sensitivity of F2R3 primers; (G) The sensitivity of F3R1 primers; (H) The sensitivity of F3R2 primers; (I) The sensitivity of F3R3 primers.













 DownLoad:
DownLoad:


