-
-
-
-
-
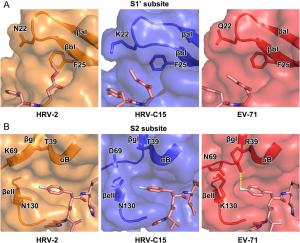
-
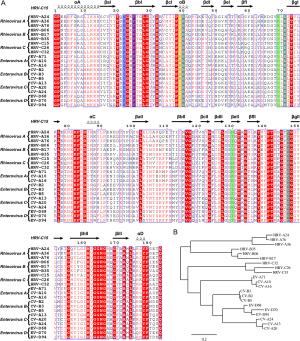
-
Name 3C 3C in complex with rupintrivir Data collection Resolution (Å) 50–2.15 (2.23–2.15) 50.00–2.05 (2.12–2.05) Unique reflections 13, 578 (1332) 16, 114 (1496) Space group C2221 C2221 Cell dimensions a (Å) 52.1 52.9 b (Å) 94.7 95.6 c (Å) 98.1 98.9 α (°) 90 90 β (°) 90 90 γ (°) 90 90 Redundancy 7.5 (6.4) 3.1 (3.0) Completeness (%) 99.8 (99.5) 92.1 (90.2) aRmerge 0.069 (0.328) 0.060 (0.263) I/σ(I) 35.6 (8.1) 22.8 (7.0) Refinement Resolution(Å) 2.15 2.05 No. reflections 13, 546 14, 812 bRwork/cRfree 0.210/0.253 0.184/0.223 No. of non-H atoms Protein 1498 1, 419 Mean B-factor (Å2) 31.5 28.0 Ramachandran statistics (%) Most favored 97.2 98.3 Allowed 2.8 1.7 Outliers 0.0 0.0 R.m.s.deviations Bond lengths (Å) 0.010 0.008 Bond angles (°) 1.089 1.086 Values in parentheses are for highest-resolution shell.
aRmerge ∑hkl∑i|I(hkl)i- < I(hkl)>|/∑hkl∑iI(hkl)i,
bRwork=∑hkl|Fo(hkl)-Fc(hkl)|/∑hklFo(hkl).
cRfree was calculated for a test set of reflections (5%) omitted from the refinement.Table 1. Data collection and refinement statistics.
Figure 6 个
Table 1 个