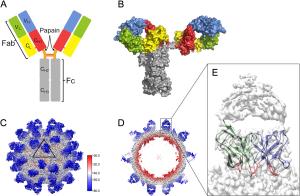
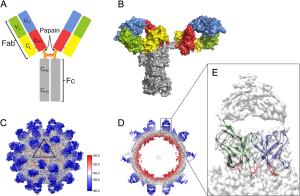
-
-
-
-
Genus Virus name Antigen Antibodya Fragment EMD codes Resolution
(Å)Enterovirus Coxsackievirus (CV) CV-A6 A-particle 1D5 Fab 6757 3.8 CV-A10 A-particle 2G8 Fab 9603 4.3 CV-A10 mature virion 2G8 Fab 9604 3.9 CV-A10 procapsid 2G8 Fab 9605 4.2 Enterovirus (EV) EV-A71 mature virion MA28-7 Fab 5673 23.4 E18 Fab 2397 10 E19 Fab 2436 13 D5 IgG 6365 7.2 D5 Fab 6366 4.8 EV-A71 procapsid D5 Fab 6383 6 22A12 Fab 6200 8.8 E18 Fab 2434 16 D6 Fab 6963 4.9 A9 Fab 6964 6.8 EV-A71 VLP D5 IgG 6384 5.5 EV-D68 mature virion 15C5 Fab 9633 3.6 15C5/11G1 Fab 9634 3.5 EV-D68 A-particle 11G1 Fab 9636 7.2 Poliovirus (PV) PV1 mature virion A12 Fab 5670 12 C3 Fab 5291 11.1 PVSP6A VHH 5886 4.8 PVSP29F VHH 5888 6.5 PVSS8A VHH 6433 4.2 PVSP19B VHH 6434 4.8 PVSS21E VHH 6435 3.8 PV2 mature virion A12 Fab 5671 20 PV1 procapsid P1 Fab 5283/5284/5285/5286 13/21/18/18 C3 Fab 5293 22 PV1 A-particle P1 Fab 5280/5282 12/26 C3 Fab 5292 9.1 PVSP17B VHH 8285 5.3 PVSS12B VHH 8285 5.3 PVSS10E VHH 8277 4.8 PVSS7A VHH 8286 5.3 Rhinovirus (RV) RV B14 mature virion C5 Fab 8754/8761/8762 2.53/2.71/2.26 RV B14 procapsid C5 Fab 8763 3.01 Aphthovirus Foot-and-mouth disease virus (FMDV) FMDV-O mature virion D9 Fab 0173 3.97 Parechovirus Human parechovirus (HPeV) HPeV-3 mature virion AT12-015 Fab 0069/3138 2.8/15 HPeV-1 mature virion AM28 Fab 2761 19.76 Hepatovirus Hepatovirus A (HAV) HAV mature virion R10 Fab 6688 4.2 F4 Fab 9827 3.9 F6 Fab 9828 3.68 F7 Fab 9829 3.05 F9 Fab 9830 3.79 aStructural insights into the possible mechanisms for antibody-mediated neutralization discussed in the text are summarized below. 1D5: inhibition of virus-cellular binding ( Xu et al. 2017 ), 2G8: capsid stabilization (Zhu R et al. 2018 ), MA28-7: cross-linking of virions and blocking receptor binding (Lee et al. 2013 ), E18: induction of genome release (Plevka et al. 2014 ), D5: capsid stabilization (Ye et al. 2016 ), 22A12: capsid stabilization (Shingler et al. 2015 ), D6: blocking receptor binding (Zhu L et al. 2018b ), A9: blocking receptor binding and capsid destabilization (Zhu L et al. 2018b ), 15C5: blocking receptor binding and locking capsid at intermediate stage, 11G1: locking capsid at intermediate stage (Zheng et al. 2019 ), R10: blocking receptor binding (Wang X et al. 2017 ), F4, F6, F7 and F9: blocking receptor binding (Cao et al. 2019 ).Table 1. Summary of cryo-EM structures from picornavirus-antibody complexes in the Electron Microscopy Data Bank (EMDB) (https://www.ebi.ac.uk/pdbe/emdb/).
-
Virus name Antigen Antibodya Fragment EMD codes Resolution (Å) Dengue virus (DENV) DENV1 mature virion 14c10 Fab 5268 7 1F4 Fab 2442 6 DENV2 mature virion 747(4)B7 Fab 2818 10.24 1A1D-2 Fab 1418 24 2D22 Fab 2967/2968/2969/2996/2997/2998/2999 6.5/20/21/6.9/13/11/23 DENV2 immature virion 2H2 Fab 5674/5675/5676/5677 21/25/21/21 E53 Fab 5102 23 DENV3 immature virion 1H10 Fab 9649/9650/9651 12/25/25 DENV3 mature virion 5J7 Fab 5935 9 West Nile Virus (WNV) immature virion E53 Fab 5103 15 Mature virion E16 Fab 1234 14.5 E16 scFv 5115 22.75 CR4354 Fab 5190 13.7 Tick-borne encephalitis virus (TBEV) Mature virion 19/1786 Fab 3754/3755 3.9/19.2 Zika virus (ZIKV) Mature virion ZIKV-117 Fab 8548 6.2 ZKA190 Fab 6793/6794 22/22 Z23 Fab 9542 9.4 C10 Fab 9573/9574/9575 4.4/12/4 ZAb-FLEP Fab 7613 9.7 ZK2B10 Fab 9811/9812 20/11 ZIKV-195 Fab 9131 4 Japanese encephalitis virus (JEV) Mature virion 2F2 Fab 6854 4.7 2H4 Fab 6855 4.6 aThe possible neutralization mechanisms for flavivirus antibodies discussed in the text are summarized below. 14c10: blocking receptor binding ( Teoh et al. 2012 ), 1F4: blocking virus attachment (Fibriansah et al. 2014 ), 1A1D-2: blocking virus attachment by binding to hidden epitopes (Lok et al. 2008 ), 2D22: blocking capsid reorganization required for virus fusion (Fibriansah et al. 2015a ), 2H2: inhibition of virus maturation (Wang et al. 2013 ), E53: binding to partially immature heterogeneous virions (Cherrier et al. 2009 ), 1H10: enhancing immature virus attachment to endosomal membrane (Wirawan et al. 2019 ), 5J7: blocking receptor binding and capsid stabilization (Fibriansah et al. 2015b ), E16: blocking capsid reorganization required for virus fusion (Kaufmann et al. 2006 ), ZIKV-117: capsid stabilization (Hasan et al. 2017 ), ZKA190: inhibition of either cell attachment or membrane fusion (Wang J et al. 2017 ), C10: capsid stabilization (Zhang et al. 2016 ).Table 2. Summary of cryo-EM structures from flavivirus-antibody complexes.
-
Genus Virus name Antigen Antibody Fragment EMD codes Resolution (Å) Lentivirus Human immunodeficiency virus (HIV) HIV-1 BaL virion A12 VHH (Tomo) 5544/5551 m36 VH (Tomo) 5552/5553/5554/5555 17b IgG (Tomo) 5456 22 VRC01 IgG (Tomo) 5457 24 VRC03 IgG (Tomo) 5458 23 VRC02 Fab (Tomo) 5459 23 VRC02 IgG (Tomo) 5460 25 VRC01/17b IgG (Tomo) 5461 28 b12 Fab (Tomo) 5018/5021 20/20 17b Fab (Tomo) 5020/5023 20/20 17b/ A32 Fab 0466 13.08 BG505 SOSIP.664 17b/8ANC195 Fab 7516 /(Tomo) 3096 3.54/23 3BNC117 Fab 8644 4.4 3BNC117/ PGT145 Fab 8643 4.3 3BC315 Fab 3067 9.3 BG1/8ANC195 Fab 8693 6.2 PG9/8ANC195 Fab 8695 11.5 3417 Fab 7552/7553/7554/7555/7556/7557 4.7/4.7/4.7/4.7/4.7/4.7 VRC34.01 Fab 8125 17 BF520.1 Fab 9166 4.8 PGT128 Fab 3121/3120 4.36/4.47 17b Fab 8730 8.6 PGV04 Fab 5779/5780/5781 5.8/7.9/8.2 PGT151 Fab 9062 4.5 BG505 DS- SOSIP.664 vFP/VRC03/PGT122 Fab 7622/7621/7459/7460 4/4/3.8/3.6 vFP Fab 8420/8421/8422 8.58/14.7/19.6 PGT145 Fab 8427 6.8 2G12/ VRC03 Fab 8981 8.8 PGT122/VRC03/ FP antibodies Fab 9189/20189/20191/9359/9320/9319/8977 3.8/4.3/3.5/3.7/4.2/4/3.18 462c SOSIP.664 VRC01GL Fab 9294/9295/9303 /9304 3.8/3.8/4.8/4.8 B41 SOSIP. 664 17b Fab 8713 3.7 PGV04 Fab 8716 7.4 b12 Fab 8717 3.6 21c/8ANC195 Fab 9038 4.06 PGT151 Fab 9030 6.7 ZM197 SOSIP. 664 VRC01 Fab 3059 9.32 PC64M18C043 FL Env PGT151 Fab 7858 3.1 PGT151/PCT64-35S Fab 7859 6.8 PC64M18C043 SOSIP. 664 PGT151 Fab 7860 4.9 PC64M4C054 SOSIP. 664 PCT64-13C Fab 7863/7864/7089 5.1/30/13.2 PCT64-13F Fab 7862 30 PCT64-35S Fab 7865/7866 5.5/8.2 PC64M4C054 FL Env PGT151/PCT64-13C Fab 7861 30 JR-FL EnvΔCT PGT151 Fab 3308/3309 4.19/4.3 PGT151/10E8 Fab 3312 8.8 AMC011 SOSIP.v4.2 PGV04 Fab 8302 6.2 KNH1144 SOSIP. gp140 VRC03 Fab 2484 6 17b Fab (Tomo) 5462 8.8 Lymphocryptovirus Epstein-Barr virus (EBV) glycoprotein AMMO1 Fab 7344/7345 4.8/10 Betacoronavirus Middle East respiratory syndrome-related coronavirus (MERS-CoV) S protein G4 Fab 8783/8784/8785/8786/8787/8788/8789/8790/8791/8792/8793 4/3.6/4.8/4.6/4.8/4.7/5/4.5/4/4/11.5 LCA60 Fab 0401/0402 3.5/3.6 Severe acute respiratory syndrome coronavirus (SARS-CoV) S protein S230 Fab 0403/0404 4.2/4.5 Alphainfluenzavirus Influenza virus Influenza virion 6F12 IgG (Tomo) 6610/6611 25/25 C179 IgG (Tomo) 5684/5685 7B2 IgG (Tomo) 6612 25 3F5 IgG (Tomo) 6613/6614 25/25 HA protein K1915 scFv 8561/8562/8563/8564 4.8/4.8/4.8/4.8 H7.5 Fab 9142/9143/9145 7.4/9.2/7.4 Ebolavirus Ebola virus (EBOV) glycoprotein 100/114 Fab 3310/3311 7.2/6.7 c2G4/c13C6 IgG/Fab 8240 4.3 c13C6/BDBV91 IgG/Fab 8241 5.5 c4G7/c13C6 IgG/Fab 8242 4.3 ADI-15878 Fab 8935/8936 4.14/4.29 VLPs c13C6 IgG (Tomo) 8226 25 c2G4 IgG (Tomo) 8227 25 c4G7 IgG (Tomo) 8228 25 Table 3. Summary of cryo-EM structures from glycoprotein–antibody complexes.
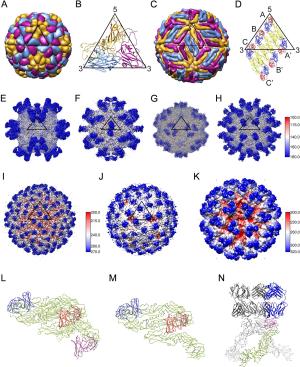
Figure 3 个
Table 3 个