HTML
-
Rabbit hemorrhagic disease virus (RHDV) is the causative agent of rabbit hemorrhagic disease (RHD), which primarily infects the wild and domestic European rabbit (Orcytolagus cuniculus) (Meyers et al. 1991b). The lesions caused by the disease are mainly in the liver, spleen, and lungs, causing acute failure with symptoms of dyspnea, disseminated intravascular coagulation, fulminant hepatic failure, and hepatocellular apoptosis (Alonso et al. 1998; Mikami et al. 1999). RHDV mainly distributes and proliferates in the liver, spleen, and lungs (Liu et al. 2015; Neimanis et al. 2018). In 2010, RHDV2, also called RHDV GI.2 (Le Pendu et al. 2017), was detected for the first time in France (Le Gall-Recule et al. 2013), spreading to other countries in Europe, Australia, Africa and America (Abrantes et al. 2013; Mahar et al. 2018; Rouco et al. 2018; Lopes et al. 2019). RHDV is a nonenveloped positive-sense single-stranded RNA virus, which belongs to the family Caliciviridae, genus Lagovirus. RHDV virions contain the genomic RNA (gRNA) and an additional 2.2 kb of subgenomic RNA (sgRNA), which is collinear with the 3′ end of the gRNA (Meyers et al. 1991a). The gRNA of RHDV consists of a positive-sense single-stranded molecule of 7, 437 nucleotides with a virus-encoded protein, VPg, which covalently attaches to its 5′ end (Zhu et al. 2015; Zhu et al. 2016). The gRNA also contains two slightly overlapping open reading frames (ORFs) of 7 kb (ORF1) and 351 nucleotides (ORF2). ORF1 is translated into a large polyprotein that is cleaved into the major structural protein VP60, the capsid protein, and seven nonstructural proteins: p16, p23, helicase, p29, VPg, protease, and RNA-dependent RNA polymerase (RdRp). RdRp is a replicase of the viral genome, and it is also prone to mutation, which can accelerate the evolution of the virus (Hukowska-Szematowicz 2020). ORF2 encodes the minor structural protein VP10 (Meyers et al. 1991a; Wirblich et al. 1995; Morales et al. 2004). The sgRNA, which only encodes VP60 and VP10, usually contributes to the production of high levels of products required during intermediate and late stages of infection (Meyers 2003). Flanking the coding regions of RHDV is a 5′-terminal noncoding region of nine nucleotides and a 3′-terminal noncoding region of 59 nucleotides (Morales et al. 2004). However, the molecular mechanisms responsible for RHDV replication remain poorly understood, mainly due to the lack of a robust cell culture system for propagation of the virus. In 2013, we developed a RHDV replicon system, which has the ability to automatically replicate in rabbit kidney cells (RK-13 cells) (Wang et al. 2013). Construction of this RHDV replicon system has provided a platform for exploring RHDV replication in host cells. In 2017, we successfully constructed mutant RHDV (mRHDV) in RK-13 cells in vitro, which has a specific receptor-recognition motif (Arg-Gly-Asp) on the surface of the capsid protein, characterized by two amino-acid substitutions. mRHDV is recognized by the intrinsic membrane receptor (integrin ɑ3β1) of RK-13 cells, by which mRHDV gains entry, replicates, and imparts apparent cytopathic effects (Zhu et al. 2017).
In this study, RHDV replicase and replicase-related host factors were isolated for the first time and its main components were identified. We found that nucleolin (NCL), a multifunctional nucleolar phosphoprotein that played a role in a variety of cell functions, was necessary for RHDV replication because the replication level of RHDV was significantly affected by knocking down the NCL gene in cells. Our data showed that NCL was a link between viral replicase and host proteins.
-
The pRHDV-luc plasmid, in which the VP60 and partial VP10 genes were replaced with the Fluc gene, was generated in our previous study (Wang et al. 2013). To generate pRHDV-luc-HA1, pRHDV-luc-HA2, pRHDV-luc-His1, pRHDV-luc-His2, pRHDV-luc-His1/HA1, pRHDV-luc-His1/HA2, pRHDV-luc-His2/HA1, and pRHDV-luc-His2/HA2 plasmids, in which the HA and His tags were inserted in-frame into RdRp, were generated by fusion PCR (Fig. 1A). The lentivirus-based expression plasmids were generated with a pLOV-CMV-GFP vector, which stored in our laboratory, using In-Fusion HD Cloning kits (No.639649, Clontech Laboratories, Inc., California, USA), according to the manufacturer's instructions. The pLOV-CMV-GFP vector was linearized with Nhe I and Not I. The plasmids, used in mammalian two-hybrid (M2H) assays, were generated with the pACT and pBIND vectors (No.E2440, Promega Corporation, Madison, USA) using In-Fusion HD Cloning kits. The p3×FLAG-CMV-14 vector (No. E7908, Sigma-Aldrich Corporation, Milwaukee, USA) and pCMV-Myc (No.635689 Clontech Laboratories, Inc.) were used to create mammalian expression constructs. All plasmids were created using In-Fusion HD Cloning kits, according to the manufacturer's instructions. In addition, the primers used in this research are listed in Supplementary Table S1.
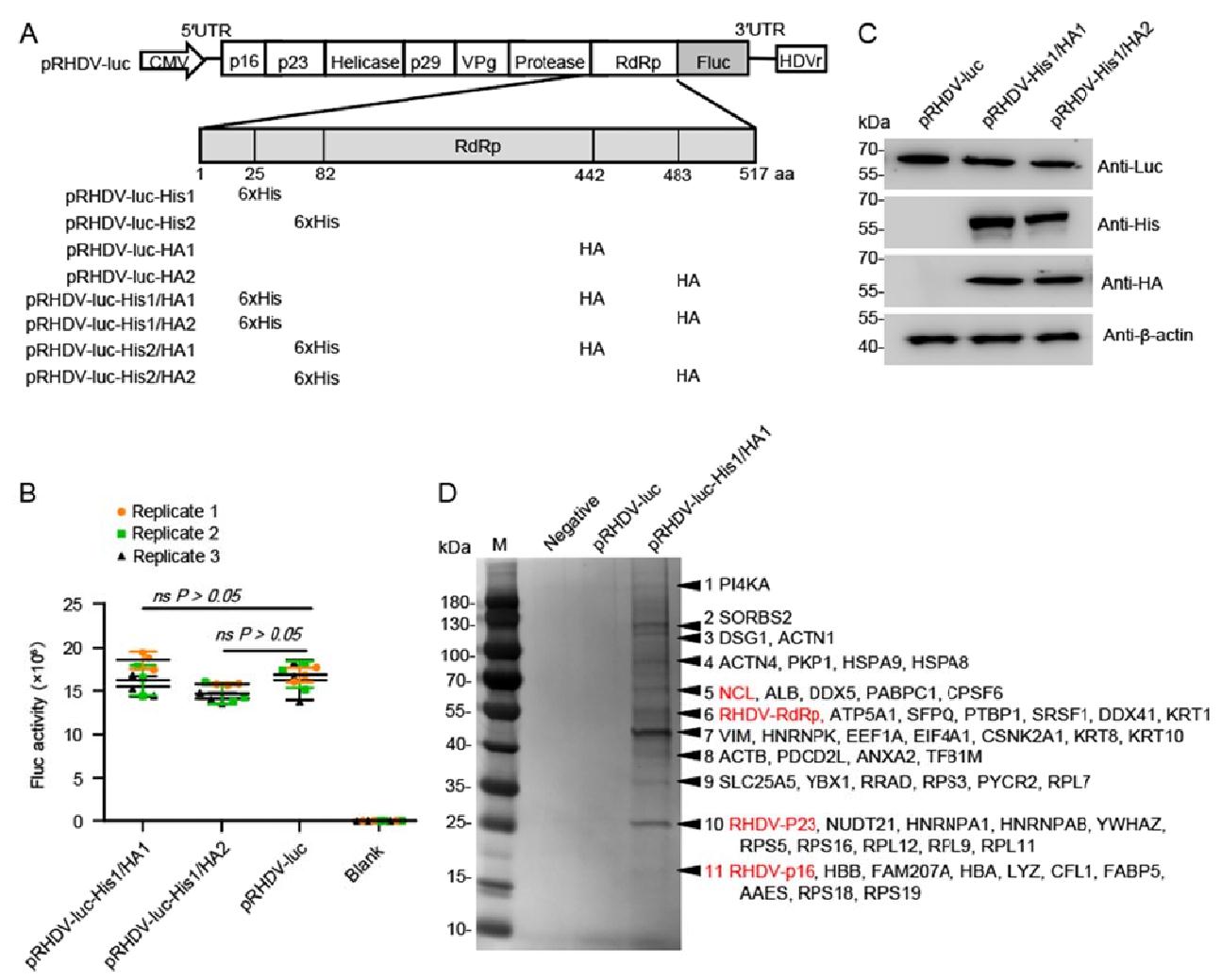
Figure 1. Affinity purification of solubilized RHDV replicase RdRp-associated host factors. A Schematic of RHDV-His/HA replicon constructs. The HA and 6×His peptide sequences are inserted into the RdRp aa sequence at 25 or 82, 442 or 483 sites, respectively. B Effect of the inserted tag on viral replicon activity. RK-13 cells were transfected with recombinant RHDV replicons. Luciferase activity in cell lysates was measured at 48 hpt. Data are shown as mean with SD. Student t-tests and analysis of variance were used for statistical analyses. Replicate 1, 2, 3 means three independent experiments, each experiment contains three technical replicate values. The number of cells used in all replicate experiments was similar. C Western blotting of recombinant RHDV replicons in RK-13 cells with the antibodies indicated. β-actin was used as an internal control. D After two-step affinity purification, the eluted proteins were resolved by SDS-PAGE. Specifically, enriched protein bands (arrows) in the pRHDV-His1/HA1-luc sample w ere identified by mass spectrometry. Mainly, host proteins and identified viral proteins in the bands indicated are shown. Host factors selected for further study are marked in red. RHDV, rabbit hemorrhagic disease virus; RdRp, RNA-dependent RNA polymerase; hpt, hours post-transfection; SD, standard deviation.
-
Rabbit kidney cells RK-13 (ATCC, CCL37) and HEK-293T cells (ATCC, CRL-3216) were routinely maintained in minimal essential medium (MEM) (No. 11090081, Gibco, New York, USA) or Dulbecco's modified Eagle's medium (DMEM) (No.11965092, Gibco), respectively, supplemented with 10% fetal bovine serum (No.04-010-1A, Biological Industries, Beit-Haemek, Israel). RK-NCL cells and RK-GFP cells were generated by transducing RK-13 cells with VSV-G pseudotyped lentiviral particles, which contained the NCL gene or GFP gene. To generate those recombinant lentiviral particles, we transfected psPAX2 (10 μg), pMD2.G (12 μg), and pLOV-NCL-GFP (22 μg), or pLOV-CMV-GFP (22 μg), respectively, into HEK-293T cells, which were seeded (1 × 106 cells) onto 100-mm tissue culture dishes, using a calcium phosphate transfection reagent (No.K278001, Invitrogen, California, USA). The lentivirus packaging and transduction procedures are based on our previous studies (Zhu et al. 2016). For the RHDV replicon replication assay, these stable cell lines, with exactly the same number of passages, were used to avoid the effect of cell passage variation on RHDV replication.
RHDV strain JX/CHA/97 was isolated in 1997 during an outbreak of RHDV in China and was stored in our laboratory. The genomic sequence of RHDV CHA/JX/97 is available in the GenBank database (accession number DQ205345). mRHDV was mutated from RHDV strain JX/CHA/97 and was stored in our laboratory (Zhu et al. 2017).
-
The antibodies used in this study included: mouse anti-His (No. ab252883), anti-HA (No. ab18181), anti-luc (No. ab185924), and anti-myc (No. ab32) antibodies purchased from Abcam (Cambridge, England); mouse anti-Flag (No. F3165) obtained from Sigma-Aldrich; mouse anti-β-actin (No. CW0096) and anti-GST (No. CW0084) antibodies purchased from Kangwei Century Biotechnology (Beijing, China); mouse anti-NCL (No. MA1-20800) and rabbit anti-NCL (No. PA5-85973) obtained from Thermo Scientific (New York, USA); mouse anti-RHDV RdRp was prepared by Genscript (Nanjing, China) and was stored in our laboratory; polyclonal rabbit anti-RHDV p16 and anti-RHDV p23 were prepared and was stored in our laboratory; goat anti-mouse IgG conjugated with HRP (No. 123-005-021) and goat anti-rabbit IgG conjugated with HRP (No. 323-005-024) were purchased from Jackson Immuno Research Europe Ltd. (West Grove, USA); goat anti-mouse IgG conjugated with Alexa Fluor 488 (No. A-10680), goat anti-rabbit IgG conjugated with Alexa Fluor 488 (No. A-11008), goat anti-mouse IgG conjugated with Alexa Fluor 633 (No. A-21050), and goat anti-rabbit IgG conjugated with Alexa Fluor 633 (No. A-21070), were obtained from Thermo Fisher Scientific. DAPI staining solution (No. C1002) was purchased from Beyotime Biotechnology (Shanghai, China). Nhe I (No. R3131S) and Not I (No. R3189S) were obtained from New England Biolabs (Ipswich, USA).
-
RK-13 cells were seeded onto ten 100-mm tissue culture dishes at a density of 1 × 106 cells/dish. The cells were grown overnight and were then transfected with pRHDV-luc-His1/HA1 (12 μg/dish) using Lipofectamine 3000 (No. L3000075, Invitrogen), according to the manufacturer's instructions. After 48 h post-transfection (hpt), the cells were washed for three times with cold Modified Dulbecco's phosphate-buffered saline (No.28344, MDPBS; Pierce™, New York, USA). The cells were then lysed with 500 μL/dish of ice-cold lysis buffer [Tris, 0.15 mol/L NaCl, 0.001 mol/L EDTA, 1% NP-40, 5% glycerol; pH 7.4, proteinase inhibitor cocktail (No.A32955, Thermo Scientific)]. After incubating cells on ice for 5 min with periodic mixing, the lysate was transferred to a microcentrifuge tube and centrifuged at ~ 13, 000 × g for 10 min to pellet the cell debris. The soluble fraction was incubated by head-to-tail rotation with 300 μL of anti-HA antibody-coated beads (No.88836, Thermo Scientific) for 4 h at 4 ℃. The beads were collected by centrifugation and were then washed for four times with 10 mL washing buffer [Tris, 0.15 mol/L NaCl, 0.001 mol/L EDTA, 1% NP-40, 5% glycerol; pH 7.4, 20 mmol/L imidazole (Thermo Scientific)]. After being washed, the bound proteins were eluted with 400 μL of wash buffer supplemented with 250 μg/mL HA peptide (No. I2149, Sigma-Aldrich) by incubation at room temperature (RT) for 10 min. After centrifugation at 3, 000 × g for 5 min, followed by a second elution with 200 μL of wash buffer supplemented with 250 μg/mL HA peptide. The eluted solutions were combined and diluted into 1.5 mL of lysis buffer containing 20 mmol/L imidazole, and 60 μL of Ni Sepharose (No.78606, Thermo Scientific) was added. After incubation at 4 ℃ for 1 h and clarification, the beads were washed for four times with 1.5 mL wash buffer. The captured proteins were eluted in 80 μL wash buffer containing 240 mmol/L imidazole and were mixed with 20 μL of 5× SDS loading buffer [250 mmol/L Tris-Cl (pH 6.8), 30% glycerol, 10% SDS, 0.02% bromophenol blue, 25% 2-mercaptoethanol)]. After being boiled for 10 min, the proteins samples were separated by SDS-PAGE, and protein bands were visualized with silver staining.
-
Total RNAs were purified using TRIzol reagent (No.15596026, Invitrogen), according to the manufacturer's instructions. DNA was removed from the isolated RNA using DNase I (No.2270A, Takara, Kyoto, Japan), and then cDNA was produced using M-MLV RT (Promega) and random hexamers. The cDNA samples were subjected to real-time PCR with SYBR Premix Ex Taq Tli RNase H Plus (No. RR820A, Takara) using an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, USA). The primers are listed in Supplementary Table S1. The relative RNA levels were determined according to the 2–∆∆CT method. The amount of mRNA in each sample was normalized to that of GAPDH.
-
The interactions between host protein and RHDV nonstructural proteins were evaluated using a CheckMate Mammalian Two-Hybrid System (No.E2440, Promega). The proteins expressed from the pACT vector recombinant plasmid acted as prey proteins, and proteins expressed from the pBIND vector recombinant plasmid acted as bait proteins. Subsequent M2H analysis was performed, according to the manufacturer's instructions. In brief, bait and prey plasmids were co-transfected with pG5luc plasmids into subconfluent HEK-293T cells at a molar ratio of 1:1:1 for pACT: pBIND: pG5luc vector. At 48 hpt, the HEK-293T cells were lysed, and Renilla luciferase (Rluc) and firefly luciferase (Fluc) activities were evaluated using the Dual-Luciferase Reporter (DLR™) Assay System (No.E1910, Promega).
-
Cells were washed with PBS and lysed in 200 μL of 1 × Passive Lysis Buffer (No. E194A, Promega). After gentle shaking for 15 min at RT, the cell lysate was transferred to a tube and centrifuged for 2 min at 12, 000 × g at 4 ºC. The supernatant (20 μL) was added to 100 μL of luciferase assay substrate to evaluate the activity of Fluc and Rluc using the Promega DLR™ assay system, based on relative light units (RLUs). Luciferase activity was analyzed using a FB12 luminometer (Berthold, Germany). To normalize the luciferase values determined for cells transfected with the Fluc replicon, Rluc activity was used as an internal control.
-
RK-13 cells were co-transfected with the bait and prey plasmids. At 48 hpt, total protein was isolated from RK-13 cells using IP lysis buffer. We conducted Co-IP analysis using a commercial Co-IP kit (No. 26149, PierceTm), according to the manufacturer's instructions. AminoLink Plus Coupling Resin was incubated with anti-myc monoclonal antibody (mAb) (ab32072, Abcam) or anti-Flag mAb (ab205606, Abcam) and then subjected to SDS-PAGE. Immunoblotting (IB) analysis of the proteins was subsequently conducted using mAbs against myc and Flag (Abcam). RNase was added to the cell lysis and wash buffers.
-
Protein samples were separated on 12% gels and were then transferred to nitrocellulose membranes (Hybond-C; Amersham Life Sciences, UK) using a semi-dry transfer apparatus (Bio-Rad Laboratories, USA). The membranes were blocked with 5% (w/v) nonfat milk in TBST buffer (150 mmol/L NaCl, 20 mmol/L Tris, and 0.1% Tween-20; pH 7.6) for 3 h at 4 ℃ and were then stained overnight at 4 ℃ with a primary antibody (Ab). After washed for three times for 10 min each, the membranes were incubated with a secondary Ab against immunoglobulin G (IgG) conjugated to horseradish peroxidase (No. A0545, Sigma-Aldrich) in PBST buffer (137 mmol/L NaCl, 2.7 mmol/L KCl, 10 mmol/L Na2HPO4, 2 mmol/L KH2PO4 and 0.1% Tween-20; pH 7.4) for 1 h at RT. Finally, after washed for three times for 10 min each, the proteins were detected using an automatic chemiluminescence imaging analysis system (Tanon Science & Technology Co., Ltd., China).
-
Cells were fixed in 3.7% paraformaldehyde in PBS (pH 7.5) at RT for 30 min and were subsequently permeabilized by incubation in methanol at −20 ℃ for 30 min. The fixed cells were blocked with 5% (w/v) nonfat milk in PBST buffer for 3 h at 4 ℃ and were then stained with a primary Ab for 2 h at 37 ℃. After washed for three times for 10 min each, the cells were incubated with a secondary Ab against IgG conjugated to fluorescein isothiocyanate (FITC) (No. F0382, Sigma-Aldrich) in PBST buffer for 1 h, at RT. Finally, after washed for three times for 10 min each, the samples were observed under a fluorescence microscope equipped with a video documentation system (ZEISS, Germany).
-
Jingjie PTM BioLab Co., Ltd. (Hangzhou, China) performed all mass spectrometry analyses. The protocol is as follows: first, gel pieces were digested with trypsin at 37 ℃ overnight. Peptides were extracted with 50% acetonitrile/5% formic acid, followed by 100% acetonitrile. Peptides were dried to completion and resuspended in 2% acetonitrile/0.1% formic acid. Second, the peptides were subjected to nano spray ion (NSI) source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to the UPLC. In order to improve the effective of mass spectrometry, the maximum injection time of peptides was set to 20 ms, and the dynamic rejection time of tandem mass spectrometry scanning was set to 15 seconds to reduce repeated scanning of precursor ions. Automatic gain control (AGC) was set at 5E4. Last, the resulting MS/MS data were processed using Proteome Discoverer 1.3 (Thermo Fisher Scientific Inc, USA). Tandem mass spectra were searched against UniProt Oryctolagus cuniculus database (https://www.uniprot.org/taxonomy/9986). Peptide confidence was set at high, and peptide ion score was set > 20.
-
Statistical analysis was performed using GraphPad Prism 6 software. Specific tests are described in the figure legends.
Plasmids
Cell Lines and Viruses
Antibodies and Chemicals
Affinity Purification of Host Factors Associated with RHDV Replicase RdRp
Quantitative Reverse Transcription (qRT)-PCR
M2H Assays
Luciferase Activity Measurements
Co-immunoprecipitation (Co-IP) Analysis
IB Analysis
Immunofluorescence Assay (IFA)
Mass Spectrometry (MS)
Statistical Analyses
-
To discover the host factors that were involved in RHDV replication, we attempted to purify the viral RHDV replicase and replicase-related host factors during viral replication and identify the associated host factors. Previously, the researchers successfully identified hepatitis C virus (HCV) replicase-associated replication complex (RC) components by inserting His and HA tags into the HCV replicon replicase NS5A and NS5B (RdRp) for affinity purification (Yi et al. 2016). Here, we aim to purify RdRp-related host factors by introducing two different tags into RdRp. We generated a recombinant replicon by simultaneously introducing His and HA tag into RdRp (position: 25 or 82 aa for His, 442 or 483 for HA, respectively) of the RHDV replicon (Fig. 1A). Fluc activity and IB analyses showed that RHDV-luc-His1/HA1 and RHDV-luc-His1/HA2 replicated similarly to the untagged RHDV replicon in RK-13 cells (P > 0.05, P > 0.05, respectively) (Fig. 1B, 1C). The SWISS-MODEL online tool (https://swissmodel.expasy.org/) predicated that insertion of the His and/or HA tag into these sites does not change the general structure of RdRp, so it is speculated that the insertion into these sites might not significantly affect RdRp activity (Supplementary Fig. S1A). Fluc activity and IB analysis showed that RHDV-luc-His1, RHDV-luc-HA1, and RHDV-luc-HA2 replicated similarly to the untagged RHDV replicon (Supplementary Fig. S1B, S1C). However, the replication level of RHDV-luc-His2 was significantly reduced. We speculate that this site has an important effect on RdRp enzyme activity, and the specific mechanism needs to be further studied.

Figure Supplementary Fig. S1 . Tagging of RHDV RdRp in the viral replicon. A RHDV RdRp protein structure analysis. For clarity, the structure of the RHDV RdRp was obtained from the Protein Data Bank (PDB) under the identification number 1khv (https://www.rcsb.org/). The structure of the RdRp mutation, as predicted by the SWISS-MODEL online tool (https://www.swissmodel.expasy.org/) and based on homology molecules found in the PDB. The orange portion indicated by the red arrow is the inserted label. B Effect of the inserted tag on viral replicon activity. RK-13 cells were transfected with recombinant RHDV replicons. Luciferase activity in cell lysates was measured at 48 h post-transfection. Statistical analysis was performed by Student t-tests. **P < 0.01. Data are shown as mean with SD. Replicate 1, 2, 3 means three independent experiments, each experiment contains three technical replicate values. The number of cells used in all replicate experiments were similar. C Western blotting of recombinant RHDV replicons in RK-13 cells with the antibodies indicated. β-actin was used as an internal control. RHDV, rabbit hemorrhagic disease virus; RdRp, RNA-dependent RNA polymerase; SD, standard deviation.
RK-13 cells were transfected with RHDV-luc-His1/HA1 replicon, which replication level is closest to the untagged replicon, and the cell lysates were sequentially purified using the HA and His tags at 48 hpt. The untagged RHDV replicon acted as a negative control. After two-step affinity purification, the eluted protein complexes were resolved by SDS-PAGE and the protein bands were visualized with silver staining. In total, 11 specific or enriched bands were sliced from the RHDV-luc-His1/HA1 lane and the proteins they contained were identified using MS (Table 1). The identified host proteins were associated with cytoskeleton components, intracellular transport, chaperone, ribonucleoprotein (RNP) components, and translation machine-related proteins. Among these proteins, numerous proteins have been shown to interact with some single-stranded positive-strand RNA viral proteins to regulate viral replication, such as HnRNPK, HSPA8, DDX5, ANXA2, and PI4KA (Hsieh et al. 1998; Saxena et al. 2012; Kovalev and Nagy 2014; Zhang et al. 2014; Dorobantu et al. 2015) (Fig. 1D and Table 1).
Category and band no. Protein score Mass (kDa) Gene name Protein description No. of unique peptides No. of peptides SC (%) Category and band no. Protein score Mass (kDa) Gene name Protein description No. of unique peptides No. of peptides SC (%) RHDV protein Cytoskeleton 6 273.3 57.8 RdRp RHDV RdRp 3 4 21.1 11 105.5 14.3 CFL1 Cofilin 1 2 6 16.8 10 186.4 25.1 p23 RHDV p23 2 3 15.5 3 316.5 102.9 ACTN1 Actinin alpha 1 2 7 32.4 11 165.8 16.2 p16 RHDV p16 2 4 10.4 4 286.3 87.4 ACTN4 Actinin alpha 4 2 6 28.5 Transport 8 357.8 41.7 ACTB Actin, cytoplasmic 1 2 5 19.0 8 57.1 39.2 ANXA2 Annexin A2 4 7 10.4 7 419.4 53.6 VIM Vimentin 13 13 25.3 1 201.3 233.6 PI4KA Phosphatidylinositol 4-kinase alpha 6 6 30.5 Chaperon 5 303.5 68.9 ALB Serum albumin 8 10 15.2 7 65.2 45.1 CSNK2A1 Casein kinase Ⅱ subunit alpha 5 5 7.5 11 287.6 15.6 HBA hemoglobin subunit alpha 5 5 40.4 4 236.2 71.0 HSPA8 Heat shock cognate 71 kDa protein 7 7 14.2 6 173.2 59.7 ATP5A1 F-type H+-transporting ATPase subunit beta 3 3 7.8 9 153.9 30.7 PYCR2 Pyrroline-5-carboxylate reductase 4 5 16.8 11 365.6 16.1 HBB Hemoglobin subunit beta 7 9 58.7 3 70.1 116.8 DSG1 Desmoglein 1 2 5 9.0 11 365.6 16.1 HBB Hemoglobin subunit beta 7 9 58.7 11 50.3 14.7 LYZ Lysozyme C 2 3 11.5 9 98.3 32.9 SLC25A5 Solute carrier family 25 3 3 11.2 4 53.5 73.5 HSPA9 Eat shock protein family A (Hsp70) member 9 2 2 13.8 11 99.8 12.6 FABP5 Fatty acid-binding protein 5 3 4 13.5 10 152.2 26.2 NUDT21 Nudix hydrolase 21 3 3 24.5 RNP complex 8 142.8 40.3 PDCD2L Programmed cell death protein 2-like 4 4 28.7 10 78.5 25.7 HNRNPA1 Heterogeneous nuclear ribonucleoprotein A1 3 7 13. 6 11 108.3 16.0 FAM207A Family with sequence similarity 207 member A 5 8 19.6 10 67.9 30.3 HNRNPAB Heterogeneous nuclear ribonucleoprotein A/B 2 6 8.9 Translation machines 7 127.2 50.9 HNRNPK Heterogeneous nuclear ribonucleoprotein K 4 9 11.6 7 94.2 50.0 EEF1A Elongation factor 1-alpha 7 7 9.3 5 87.0 67.5 DDX5 DEAD-box helicase 5 2 4 8.3 11 135.4 17.7 RPS18 Ribosomal protein S18 4 4 18.7 5 90.6 69.4 NCL Nucleolin 8 10 28.8 11 113.3 16.3 AAES 40S ribosomal protein S14 3 3 28.3 2 117.8 130.7 SORBS2 Sorbin and SH3 domain containing 2 5 5 14.7 10 174.3 20.2 RPL11 Ribosomal protein L11 4 4 20.1 10 135.6 26.2 NUDT21 cleavage and polyadenylation specificity factor subunit 5 3 3 10.4 10 283.1 21.2 RPL12 Ribosomal protein L12 5 5 33.5 5 99.1 70.9 PABPC1 Polyadenylate-binding protein 1 4 4 16.3 11 164.9 12.8 RPL30 60S ribosomal protein L30 4 4 50.5 9 128.4 36.0 YBX1 Nuclease sensitive element-binding protein 1 5 6 22.1 11 68.3 15.5 RPS17 40S ribosomal protein S17 3 3 29.6 6 112.1 57.4 PTBP1 Polypyrimidine tract-binding protein 1 4 7 6.7 10 323.2 22.9 RPS5 Ribosomal protein S5 7 7 18.5 6 65.2 52.1 SRSF1 Splicing factor, arginine/serine-rich 1 6 7 8.8 9 258.2 31.1 RPS3 40S ribosomal protein S3 9 9 25.8 5 75.9 63.5 CPSF6 Cleavage and polyadenylation specificity factor subunit 6/7 3 10 10.9 7 74.4 45.3 EIF4A1 Eukaryotic initiation factor 4A-I 4 6 9.7 6 75.2 62.5 DDX41 Probable ATP-dependent RNA helicase 3 12 14.5 11 122.6 13.7 RPS25 Ribosomal protein S25 5 5 21.3 11 75.3 14.0 RPS15A Ribosomal protein S15a 2 5 18.3 a Protein lists for each of the proteins identified in Fig. 2B. SC (%) refers to the percent sequence coverage for the protein. Table 1. Categories of host factors found to be associated with RHDV replicasea.
-
NCL is a phosphoprotein that is ubiquitously and abundantly expressed in many eukaryotic cells and is highly conserved during evolution, as it is involved in a remarkably large number of cellular activities (Jia et al. 2017). It has been confirmed that NCL also plays important roles in the replication and intracellular trafficking of multiple viruses (Tuteja and Tuteja 1998; Bicknell et al. 2005; Hirano et al. 2005; Becherel et al. 2006; Mongelard and Bouvet 2007; Abdelmohsen and Gorospe 2012; Durut and Saez-Vasquez 2015). To determine if NCL was required for RHDV replication, RK-13 cells were co-transfected with an RHDV replicon, NCL siRNA or Flag-tagged NCL plasmids and internal control plasmid encoding an Rluc gene. The reporter luciferase activity was evaluated using a dual-luciferase reporter assay system with cell lysates that were harvested at 24 and 48 hpt. Fluc activity was normalized with respect to a co-transfected plasmid encoding an Rluc. Similar results were obtained in three independent experiments. The results showed that there is a positive correlation between the expression level of Fluc and NCL. The Fluc activity decreased with increasing NCL siRNA transfection dose and increased with increased dose of Flag-NCL transfection (Fig. 2A, 2B). In previous study, we found that the RHDV replicon activity reached a maximum value at 24 hpt and then declined obviously at 48 hpt (Wang et al. 2013). In this study, it was found that after overexpression of NCL, the replication level of RHDV replicon at 48 hpt is close to the 24 hpt level. Therefore, the expression of NCL promotes RHDV replication in a dose-dependent manner. Subsequently, we examined the effect of NCL on mRHDV, which could proliferate in RK-13 cells (Zhu et al. 2017). We also used NCL siRNA or Flag-tagged NCL to change the expression level of NCL, and then infected the cell with mRHDV (MOI = 1). At 48 h post-infection (hpi), the replication level of mRHDV was detected by Western blotting (WB) and qPCR. The results were similar to RHDV replicons. As shown in Fig. 2C and 2D, the replication level of mRHDV increased with increased dose of Flag-NCL and decreased with increasing NCL siRNA. In addition, we successfully constructed an RK-NCL cell line, which overexpressed the NCL gene, using a lentiviral packaging system (Fig. 2E). To evaluate the replication of RHDV in RK-NCL cells, the cells were co-transfected with an RHDV replicon and internal control plasmid encoding an Rluc gene. The reporter luciferase activity was evaluated at 24 hpi. The results showed that the expression level of Fluc in RK-NCL cells was significantly higher than that in control cells (RK-GFP cells and RK-13 cells) (Fig. 2F). Collectively, these data suggest that NCL is involved in RHDV replication.
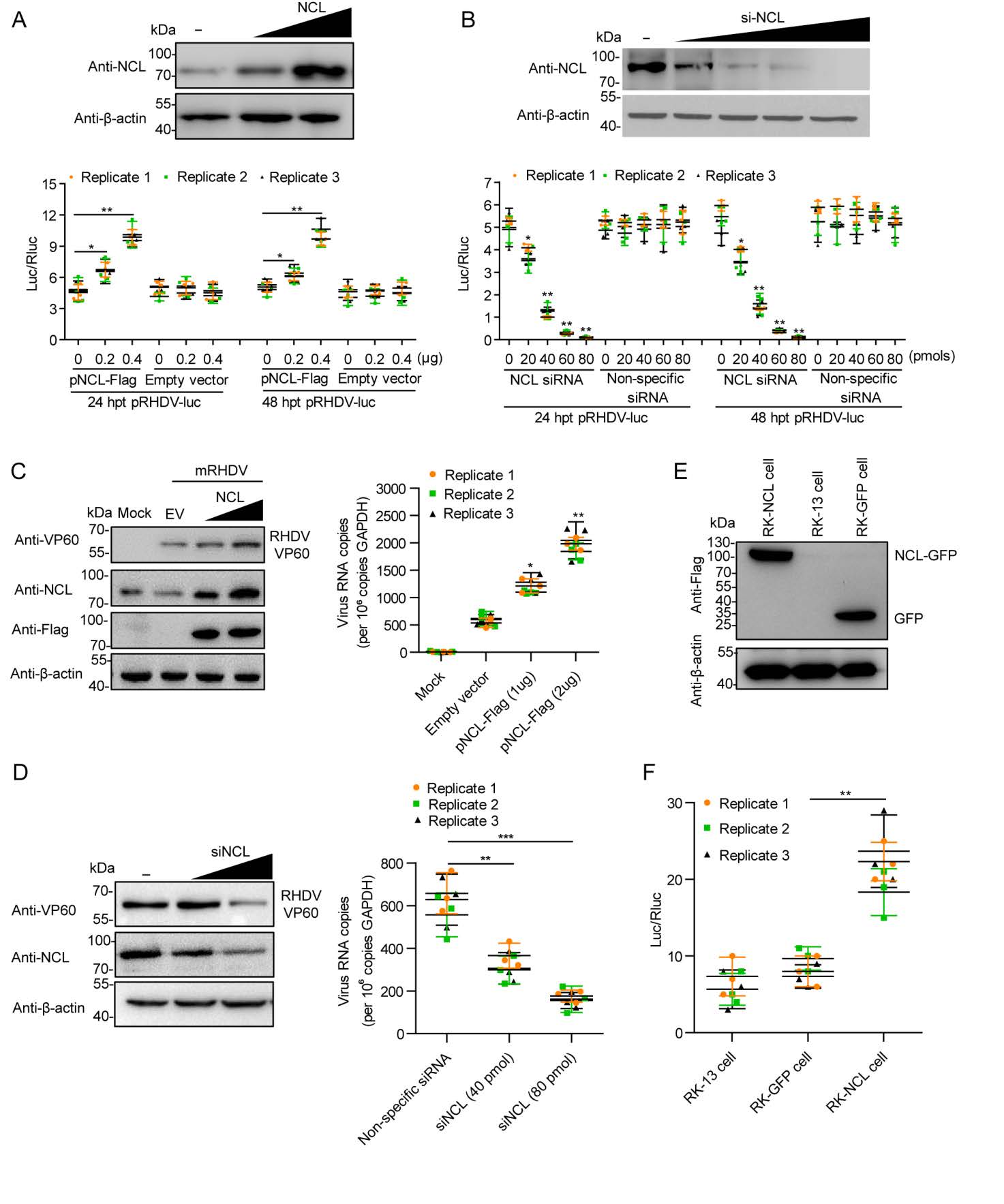
Figure 2. NCL is involved in RHDV replication. A The effect of NCL eukaryotic plasmids on viral replicon activity. Relative luciferase activity was evaluated in RK-13 cells carrying pRHDV-luc, and trans-supplemented NCL eukaryotic plasmids pFlag-NCL (0.2 μg, 0.4 μg) at 24 h post-transfection (hpt) and 48 hpt. The p3×Flag-CMV-14 vector acted as negative control (–). The luciferase activity in RK-13 cells was evaluated by measuring Fluc activity. Rluc activity was measured to normalize the transfection efficiency. B The effect of NCL siRNA on viral replicon activity. The RK-13 cells, co-transfected with pRHDV-luc and NCL siRNA (20 pmol, 40 pmol, 60 pmol, or 80 pmol), were lysed at 24 hpt and 48 hpt, and Fluc activity was measured based on RLUs and normalized according to the results obtained for a co-transfected pLTK plasmid encoding Rluc. The nonspecific siRNA acted as negative control. C–D The effect of NCL on mRHDV replication. The RK-13 cells, transfected with pFlag-NCL (1 μg, 2 μg) or NCL siRNA (40 pmol, 80 pmol), were infected with mRHDV (MOI = 1) at 24 hpt, and the level of mRHDV replication were evaluated by Western blotting and qRT-PCR at 48 hpi. The p3×Flag-CMV-14 vector (EV) and nonspecific siRNA (–) acted as negative control. RK-13 cell acted as mock control. E The expression level of NCL in RK-NCL cells at 10 passages was determined by Western blotting analysis with anti-Flag mAb. F The RHDV replicon replication levels in RK-NCL cells were evaluated by measure Luc at 24 hpi. RK-GFP cells acted as negative controls; RK-13 cells acted as blank controls. Statistical analysis was performed by Student t-tests. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean with SD. Replicate 1, 2, 3 means three independent experiments, each experiment contains three technical replicate values. The number of cells used in all replicate experiments was similar. RHDV, rabbit hemorrhagic disease virus; NCL, nucleolin; hpt, hours post-transfection; SD, standard deviation.
-
To determine if NCL regulates RHDV replication through interaction with viral nonstructural proteins, we used M2H assays to screen the interaction between NCL and viral nonstructural proteins. As shown in Fig. 3A, NCL interacted with RdRp, p16, and p23. Moreover, an IFA was performed using NCL mAbs, RdRp mAbs, p16 polyclonal antibody and p23 polyclonal antibody in RK-13 cells infected with mRHDV at 24 hpi. As shown in Fig. 3B, NCL was co-localized with RHDV RdRp, p16 and p23 in the RK-13 cell cytoplasm. In addition, the distribution of NCL in the cytoplasm increased after RHDV infection. To determine whether endogenous NCL binds to these viral nonstructural proteins, during RHDV genome replication, we assessed the interaction between NCL and these viral proteins in RK-13 cells, in the presence and absence of mRHDV infection for 24 h at 37 ℃. The results of an IP assay performed with cell lysates using NCL mAb showed that regardless of whether the cell lysates were treated with RNase or not, NCL interacted with RdRp, p16, and p23 in infected cells, but did not in uninfected cells (Fig. 3C). To prove NCL interacts with RdRp, p16, and p23, a series of Co-IP assays were used with a myc mAb in RK-13 cells, which were co-transfected with pRdRp-myc, p16-myc, p23-myc and pNCL-Flag eukaryotic expression plasmids. We showed that overexpressed NCL-Flag was present in the anti-myc immunocomplex (Fig. 3D). These results showed that NCL could interact with RHDV RdRp, p16, and p23.
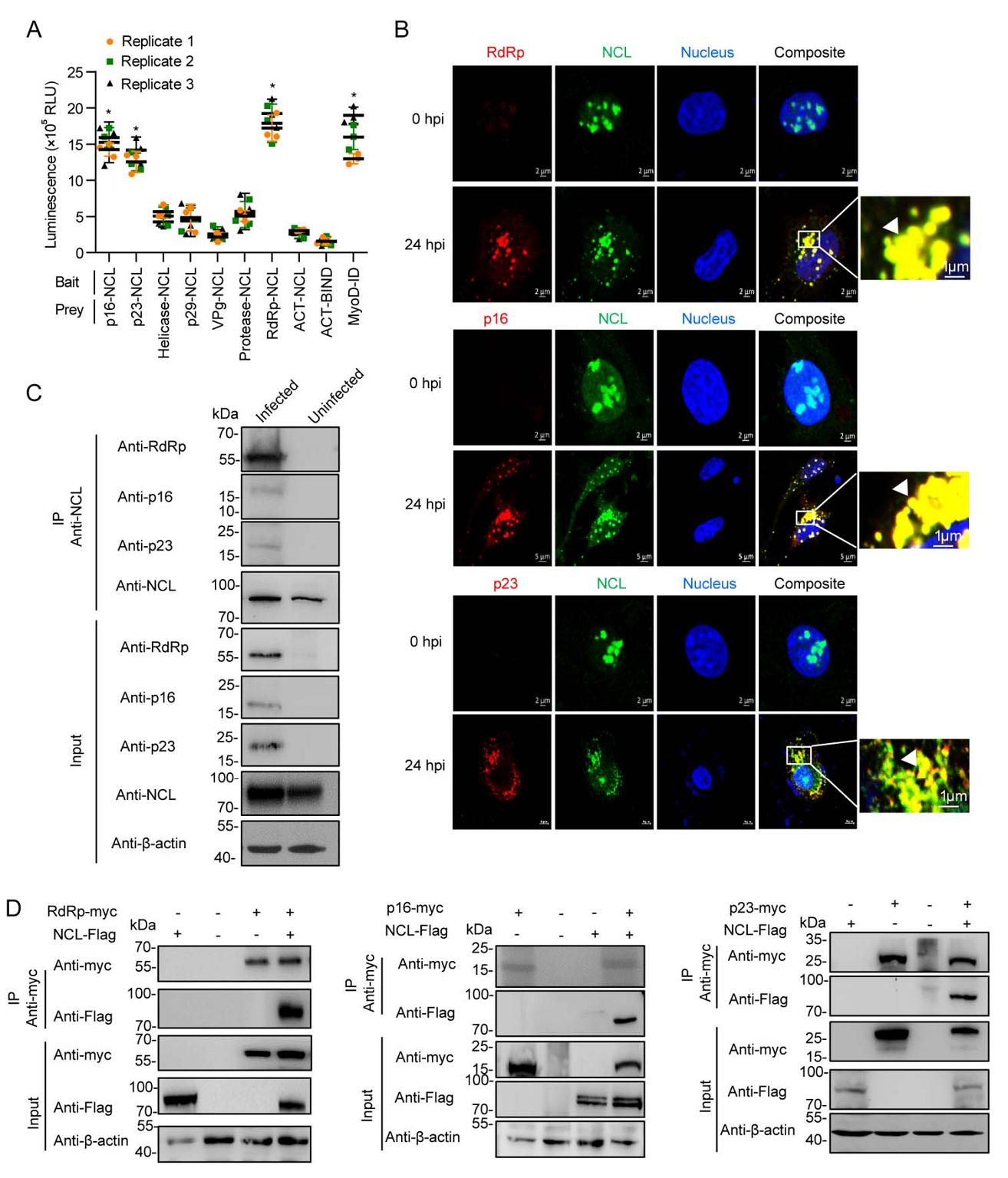
Figure 3. NCL interacts with RHDV replicase RdRp, nonstructural proteins p16 and p23. A M2H interaction of NCL with RHDV nonstructural proteins. Statistical analysis was performed by Student t-tests. *P < 0.05. Data are shown as mean with SD. Replicate 1, 2, 3 means three independent experiments, each experiment contains three technical replicate values. The number of cells used in all replicate experiments was similar. B Confocal microscopy analysis of NCL (green), RdRp (red), p16 (red) and p23 (red) in RK-13 cells infected with mRHDV at 24 h post-infection with mAbs against NCL and RdRp. The small white boxes represent amplified random co-localization spots within the merged image, and the co-localization spots are indicated with white arrowheads. C NCL binds to RdRp, p16 and p23 during RHDV replication. An IP assay was performed on cell lysates using NCL mAb in RK-13 cells that were infected or uninfected with mRHDV, then immunoblotted with Abs against NCL, RdRp, p16 or p23. β-actin was used as an internal control. Cells uninfected with mRHDV served as negative controls. D Validation of the interaction of RHDV RdRp, p16 and p23 with NCL in a Co-IP assay. RK-13 cells were co-transfected with the indicated plasmids (+) or empty vectors (–). At 48 h post-transfection, cells were lysed, and IP of myc-fused proteins was performed using anti-myc mAb. Lysates (input) and IPs were analyzed with IB using antibody against myc or Flag. β-actin was used as an internal control. RHDV, rabbit hemorrhagic disease virus; RdRp, RNA-dependent RNA polymerase; SD, standard deviation; NCL, nucleolin; IP, immunoprecipitation; IB, immunoblotting; mAb, monoclonal antibody.
-
The positive-strand RNA viruses share a conserved replication mechanism in which viral proteins induce host membrane modification to assemble membrane-associated viral replication complex (RCs) (den Boon and Ahlquist 2010). Viruses hijack host factors to facilitate this energy-unfavorable process (Nagy and Pogany 2011). Therefore, the components of the viral RC are numerous and complex. NCL is capable of binding to nonstructural proteins (RdRp, p16, and p23) of RHDV. To test the hypothesis that NCL acted as a platform for the RdRp to be attracted to the p16 and p23 proteins, a series of HA tag affinity purification analyses were performed. RHDV-luc-His1/HA1 replicon was co-transfected with NCL siRNA, or non-specific siRNA, in RK-13 cells, respectively, or was transfected in RK-NCL cells. Using IB to detect the purified RdRp-associated protein, we found that the RdRp-associated protein content was significantly reduced in NCL siRNA-treated cells and significantly increased in RK-NCL cells (Fig. 4A). To investigate the specific role of NCL in the formation of RHDV RCs, M2H assays were used to screen the interactions between viral nonstructural proteins and multiple host factors in RCs. As shown in Fig. 4B, there are complex interactions between viral nonstructural proteins and host factors in RCs. For example, p16 interacts with itself, helicase, p29, protease, and NCL; p23 binds to protease and NCL; p29 binds to helicase and VPg; helicase interacts with itself; VPg binds to protease; protease interacts with itself and RPS5; RdRp interacts with RPS5; and NCL binds to HnRNPK, CSNK2A1, RPS5, and RPL11. We subsequently used a series of Co-IP assays with a myc mAb in RK-13 cells, which were co-transfected with bait (myc fusion protein) and prey (Flag fusion protein) eukaryotic expression plasmids. IB analysis using a mAb against Flag showed the specific band corresponding to prey proteins in the myc Co-IP assay (Fig. 4C). These results reveal that RHDV replicase RdRp cannot directly bind to other nonstructural proteins of the virus. It is noteworthy that NCL directly interacts with RHDV RdRp and nonstructural proteins (p16 and p23). Together, these data suggest that RHDV completes its replication process by hijacking NCL to recruit other viral proteins and host factors.
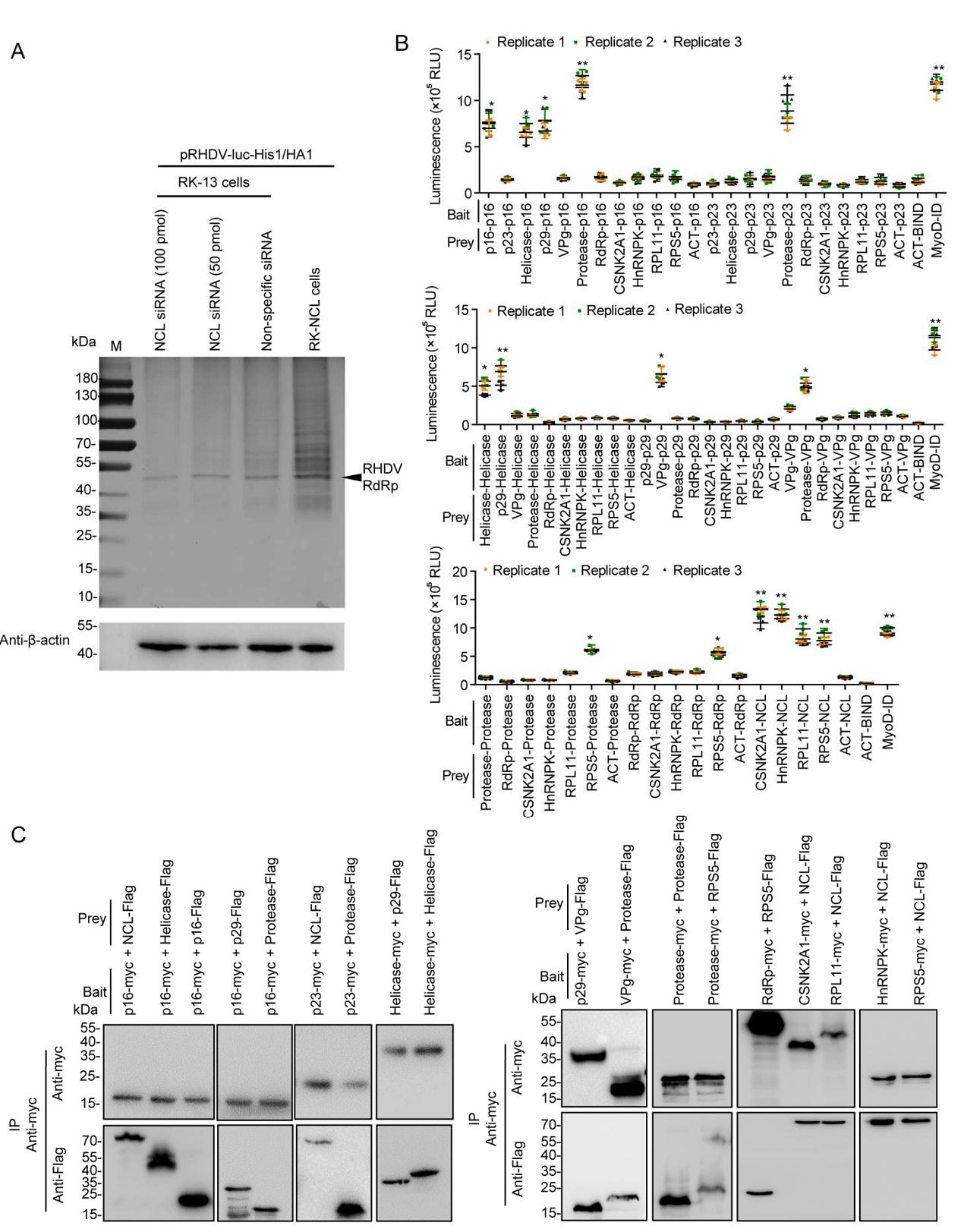
Figure 4. Identification of interactions between RHDV nonstructural proteins and host factors of RCs. A NCL siRNA inhibited the formation of the RHDV RC. After HA tag affinity purification, the eluted proteins were resolved by SDS-PAGE. The protein bands were visualized with silver staining. PBS acted as a negative control; β-actin acted as an internal control and was detected by IB with mAb against β-actin. B Identification of these interactions by M2H assays. Bait and prey plasmids were co-transfected with pG5luc plasmids into subconfluent HEK-293T cells at a molar ratio of 1:1:1 for the pACT: pBIND: pG5luc vector. At 48 h post-transfection (hpt), the HEK-293T cells were lysed, and Rluc and Fluc activities were evaluated using the Promega Dual-Luciferase Reporter Assay System. All experimental groups were compared with the negative control group (ACT-Bind). Statistical analysis was performed by Student t-tests. *P < 0.05 and **P < 0.01. Data are shown as mean with SD. Replicate 1, 2, 3 means three independent experiments, each experiment contains three technical replicate values. The number of cells used in all replicate experiments was similar. C These interactions were verified using Co-IP assays. RK-13 cells were co-transfected with bait and prey plasmids. Cell lysates were prepared 48 hpt and the proteins were subjected to IP followed by IB analysis. myc fusion proteins acted as bait proteins and Flag fusion proteins acted as prey proteins. RHDV, rabbit hemorrhagic disease virus; RC, replication complex; IP, immunoprecipitation; IB, immunoblotting; mAb, monoclonal antibody; SD, standard deviation.
Identification of Host Factors Associated with RHDV Replicase RdRp
NCL is Involved in RHDV Replication
NCL Interaction with RHDV Replicase RdRp, Nonstructural Proteins p16 and p23
NCL is a Link between RHDV Replicase and Host Proteins
-
As an important member of the positive-strand RNA viruses, the Caliciviridae family has attracted increasing attention because it contains many viruses that infect a wide spectrum of hosts and are a growing threat to human and animal health. However, most caliciviruses cannot be cultured in vitro, including some important pathogens such as RHDV and Norovirus (NV); therefore, the replication and pathogenic mechanisms of these viruses remain poorly understood. The emergence and advancement of reverse genetic manipulation technology has provided an excellent operating platform for revealing the molecular mechanism of calicivirus replication. For example, using a NV replicon, a series of works have been carried out to reveal the mechanism of NV replication (Subba-Reddy et al. 2012; Thackray et al. 2012; Thorne and Goodfellow 2014; Li et al. 2018; Oliveira et al. 2018). Recently, we also successfully established an RHDV replicon operating platform (Wang et al. 2013) and have applied it to study the genomic structure and function of RHDV.
In this study, we purified viral replicase and identified the replicase-associated host factors using an RHDV replicon system in which two different affinity tags were simultaneously inserted in-frame into RdRp. We determined that NCL played a key role in the formation of RHDV RCs. On the one hand, NCL binds to RHDV replicase (RdRp) (Fig. 3). Similar to other positive-sense RNA viruses, RHDV RdRp has a central role in the viral replication cycle. RdRp has many enzymatic properties; it binds template RNAs, initiates replication, catalyzes elongation, and terminates replication (Urakova et al. 2017b). Moreover, RdRp is able to induce the redistribution of Golgi membranes in kidney and liver cell lines of three different species (Urakova Nadya et al. 2017a). In addition, previous studies have also shown that a certain percentage of RHDV RdRp enters the nucleus, but its role is still unclear (Urakova et al. 2015). On the other hand, NCL also interacts with some important host factors, such as HNRNPK, RPL11, CSNK2A1, and RPS5 (Fig. 1, 4). Previous studies have shown that these proteins are involved in the replication of various viruses. For example, HNRNPK has been reported to recognize the 5′-terminal sequence of HCV RNA (Fan et al. 2014); CSNK2A1 interaction with NS1 plays an important part in parvovirus replication (Nüesch and Rommelaere 2006); and RPL11 and RPS5 are components of the ribonucleoprotein complex (Maggi et al. 2008; Robledo et al. 2008). Therefore, we hypothesize that these host factors may also be involved in the replication of RHDV.
We also identified that NCL interacts with nonstructural proteins (p16 and p23) of RHDV (Fig. 3). Previous studies have reported that p16 can accumulate in subnuclear compartments, which may point to a specific interaction with nucleic acids and/or cellular proteins (Urakova et al. 2015). A similar nuclear/subnuclear accumulation of nonstructural proteins has been reported in picornavirus, for which nuclear accumulation of the 2A protein and a close association of this protein with the nucleolar ribosomal chaperone protein B23 have been reported. It was suggested that 2A upregulated the formation of modified ribosomes with a preference for viral internal ribosomal entry sites, thereby contributing to the inhibition of cap-dependent cellular mRNA translation (Aminev et al. 2003). The N-terminal domain of NCL contains acidic regions, rich in glutamic acid and aspartic acid, which are the sites of phosphorylation and participate in the transcription of rRNA and interact with components of the pre-rRNA processing complex (Jia et al. 2017). Therefore, we speculate that RHDV utilizes the interaction between p16 and NCL to hijack NCL-associated machinery of rRNA transcription and pre-rRNA processing to replicate viral gRNA. In addition, the role of p23 in RHDV replication is still unclear. Previous studies have found that, similar to other caliciviruses, RHDV p23 is enriched in the endoplasmic reticulum membrane and plays an important part in inducing intracellular membrane rearrangement (Urakova et al. 2015). Therefore, we believe that p23 interacts with NCL to recruit replication-associated host proteins to the endoplasmic reticulum membrane and assembles these to form membrane-associated RCs.
NCL is an abundant and ubiquitously expressed protein in many growing eukaryotic cells (Jia et al. 2017). NCL is mainly localized within the nucleolus, but it also exists in the nucleoplasm, cytoplasm, and cell surface (Tuteja and Tuteja 1998; Abdelmohsen and Gorospe 2012). NCL controls a wide range of fundamental cellular processes, such as ribosome biogenesis, proliferation, and cell cycle regulation, and it also has important roles in the infection process of multiple viruses (Jia et al. 2017). For example, NCL acts as a receptor for human respiratory syncytial virus (Tayyari et al. 2011). NCL also mediates cellular attachment and internalization of enterovirus 71 (Su et al. 2015). A recent study showed that NCL interacted with the capsid protein of dengue virus, suggesting a role in viral morphogenesis (Balinsky et al. 2013). In previous studies, we also found that NCL mediated the internalization of RHDV through interacting with the RHDV capsid protein (Zhu et al. 2018). Of note, NCL also plays an important part in replication of several viruses. For example, NCL interacts with the feline calicivirus (FCV) NS6 (protease) and NS7 (polymerase) proteins, and has a role in virus replication (Cancio-Lonches et al. 2011). Similarly, the interaction between NCL and the UTRs of FCV (Cancio-Lonches et al. 2011) and poliovirus (Waggoner and Sarnow 1998) stimulates translation of viral proteins. Moreover, NCL binds to a protein of herpes simplex virus type 1 to facilitate the export of US11 from the cell nucleus to the cytoplasm (Sagou et al. 2010). However, the function of NCL in binding to RHDV replicase and recruiting host factors to form RCs had not been revealed until now. Here, we demonstrate for the first time that NCL can specifically bind to RHDV replicase (RdRp) and can act as a link, recruiting host factors and viral proteins, to form RHDV RCs. Our findings enrich the current knowledge about the mechanism of NCL regulation of viral replication and provide new clues for further exploration of the interaction between RHDV and host proteins.
In conclusion, we identified the host factors associated with RHDV replicase, for the first time. We found that NCL acted as a link to recruit host factors, viral replicase (RdRp), and nonstructural proteins (p16 and p23), thereby forming a complex and ordered RHDV RC. Elucidation of the molecular mechanism by which NCL regulates viral replicase assembly may lead to new insights into viral RC biogenesis and novel antiviral strategies.
-
This study was sponsored by Shanghai Sailing Program (20YF1457700), the National Natural Science Foundation of China (32000109 and 31672572), and the China Postdoctoral Science Foundation (2019M660885 and 2021T140718). In addition, we thank LetPub for its linguistic assistance during the preparation of this manuscript.
-
GL, GT, JZ, QM, HG and RQ conceived and performed the experiments. JZ, AT, DD, JT, and FW analysed the data. JZ prepared the figures. JZ and QM wrote the original draft. GL and GT checked the finalized manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.













 DownLoad:
DownLoad:




