-
Baculovirus produces two types of viral progeny with distinctly different structural and biological properties: the budded virus (BV) and the occlusionderived virus (ODV). ODVs and BVs, although their nucleocapsids are similar, are morphological distinct and have specific polypeptides and have different functional roles: ODVs, responsible for oral infection, are highly infectious for midgut epithelial cells; BVs spread disseminate the virus in susceptible larval tissues causing a secondary infection and are highly infectious to cultured cells. The two viral forms are produced in a sequential manner in the same cells: BVs are produced in the early stages, allowing the virus to spread to all susceptible larval tissues. Later, the virions are retained within the nuclei where they acquire specific proteins and are eventually embedded into the occlusion body (OB) (1, 15).
The life cycle of baculoviruses in nature starts when larvae ingest of OBs present on contaminated foliage. The OBs are dissolved in the midgut and the contained virions (ODVs) are released, pass through the peritrophic membrane and infect midgut cells. Until now, four genes, p74, pif-1, pif-2 and pif-3, have been shown to be involved in per os infection. P74 is a structural protein of ODV in Autographa californica multicapsid nucleopolyhedrovirus (Ac-MNPV) and necessary for infectivity of polyhedra (5, 11), which might function to bind to a specific receptor on target cells (7). PIF-1 and PIF-2 were first identified as essential factors for per os infectivity of ODV in Spodoptera littoralis NPV and S. exigua NPV, respectively (10, 14). In AcMNPV, PIF-1 (Ac119), PIF-2 (Ac022) and PIF-3 (Ac115) have also been identified as essential factors for oral infection of ODV (13). PIF1 and PIF2 as ODV envelope attachment proteins mediate specific binding to primary target cells within the midgut. In contrast, PIF 3 mediates another uniden-tified, but critical, early event during primary infection (13).
Although some insights have been gained in recent years, the molecular mechanism of oral infection is still unclear, mainly due to the lack of an in vitro system. Horton and Burand reported that Lymantria dispar NPV (LdMNPV) ODVs could infect cultured L. dispar cells (IPBL-LdEIta) (9), but the mechanism has not been analyzed in detail.
Helicoverpa armigera nucleopolyhedrovirus (HearNPV) has been widely used to control the host in China. Sequence analysis indicates that the genome is about 130kb containing 135 ORFs (2). The homologues of p74, pif-1, pif-2 and pif-3 were encoded by HearNPV ORF20, ORF 111, ORF 132 and ORF 98, of which ORF132 (pif-2) has been identified as an essential factor for per os infectivity (4, 17). In this report, we investigated if ODV could infect cultured cell line. Results demonstrated that HearNPV ODV could infected Hz-AM1 cells in pH dependent way. The optimal pH for infection was 8.5, which is same to that in the microvilli of midgut epithelial cells, the ODV native infection sites. Antibodies neutralization analysis indicated that four HearNPV oral infection essential genes p74, pif-1, pif-2 and pif-3 were also essential for HearNPV ODV infection in vitro. Thus, HearNPV–HzAM1 system can be used to analyze the mechanism of ODV entry.
HTML
-
The H. zea cell line (BCIRL-Hz-AM1, Hz-AM1) was used and maintained at 28℃ in Grace's medium supplemented with 10% fetal bovine serum (FBS) (12). HearNPV G4 and the recombinant HaCXW1, which contains a gfp gene under control of polh promoter in the egt locus (2, 3) were used for analysis.
-
HearNPV and HaCXW1 polyhedra were amplified in H armigera larvae. ODVs were isolated from polyhedra according to the previously described methods (2). The ODV pellet was resuspended in phosphate-buffered saline (PBS; 150 mmol/L NaCl, 10 mmol/L phosphate buffer, pH 7.4). Protein determinations were used to calculate virus particles according to the previously described. It is about 1.2×109 particles per μg of ODV protein (16).
-
The truncated p74 (from 1 to 1332 bp of the ORF), pif-1 (ha111) and pif-3 (ha98) were amplified from HearNPV G4 genome by PCR with certain primers (Table 1), then cloned into the expression vector pET28a+ or pGEX-KG, and generated the recombinant plasmids pET28a-p74, pET28a-pif1 and pKGpif3, in which the target gene would generate a protein fused to the C-terminal of the tag. Escherichia coli DH5α cells containing each plasmid were grown to an OD600 of 0.4 and then induced with 1 mmol/L IPTG, respectively. After 3 h at 37℃, cells were harvested and lysed with lysozyme, sonicated, and centrifuged at 5 000×g for 10 min at 4℃. The fusion proteins, present in the pellet were separated in 12% SDS polyacrylamide gels and purified. Antisera (αHap74, αHaPIF1 and αHaPIF3) were generated by immunizing rabbits with purified proteins and tested by Western blot analysis. Antisera were neutralized with both E. coli and HzAM1 lysate. The antiserum to HearNPV PIF-2 (Ha132) (αHa132) was kindly provided by Dr. Zhihong Hu (4).

Table 1. The primers used in the experiments
-
1×105 Hz-AM1 cells were seeded into every well of 24-well tissue culture plate and incubated for 12-18 h. The cells were washed three times with PBS (pH7.2), and then were infected with 1.19×108 ODV, which diluted in PBS with different pH values (7.2, 8.0, 8.5, 9.0, 9.5). The viruses were allowed to adsorb for 1 h at 28℃, then removed and incubated at 28℃ in Grace's medium supplemented with 10% FBS. Cells expressing GFP were detected and calculated by fluorescence microscopy at 72 h post infection (hpi).
-
The purified ODVs(1.19×108) were diluted with 200 μL PBS (pH7.2) containing 0.5% of either antiserum or normal rabbit serum, and incubated for 1 h at 37℃. The neutralized and mock-neutralized viral preparations were added to 1×105 cells per well of a 24-well tissue culture plate for 1h at 28℃ (8), then removed and incubated at 28℃ in Grace's medium supplemented with 10% FBS. Cells expressin GFP were detected and calculated by fluorescence microscopy at 72 hpi.
-
Each figure displays representative results from three independent experiments. All experimental data were calculated from triplicate samples. Data were analyzed using independent sample t-tests and are expressed as means ± standard deviation (SD) and p-values less than 0.05 were considered significant.
Cells and viruses
Preparation of the ODVs
Preparation of antiserum against HearNPV PIFs
ODVs infection in Hz-AM1 cells
Virus neutralizations
Statistical analysis
-
The fragments of p74, pif-1 and pif-3 were amplified by PCR and inserted into pET28a+ or pGEXKG vectors, respectively. Those expression plasmids were named pET28a-p74、pET28a-pif1 and pKG-pif3, which were confirmed by restriction enzyme analysis and sequencing. The fusion proteins were then expressed in E. coli, and were separated in 12% SDS polyacrylamide gels. The size of P74-His, PIF1-His and PIF3-GST fusion protein was about 49 kDa, 55 kDa and 48.5 kDa, respectively (Fig. 1A), which is similar to what we expected. Antiserums (αP74, αPIF-1 and αPIF-3) were generated by immunizing rabbits with purified proteins in rabbits and could specifically recognize 49 kD, 55 kD and 48.5 kD bands (Fig. 1B).

Figure 1. Expression analysis of pET28a-p74, pET28a-Ha111 and pKG-Ha98 (A). M, Protein marker; 1, Induced expression of pET28a-Ha111; 2, Uninduced expression of pET28a-Ha111; 3, Induced expression of pET28a-p74; 4, Uninduced expression of pET28a-p74; 5, Uninduced expression of pKG-Ha98; 6, Induced expression of pKG-Ha98. The arrows indicate the recombinant fusion proteins. Identification of antisera using Western-blot analysis (B). M, Prestained protein marker; 1, 3, 5, Control using normal serum; 2, Ha111 protein is recognized with αHa111; 4, P74 protein is recognized with αp74; 6, P98 protein is recognized with αp98.
-
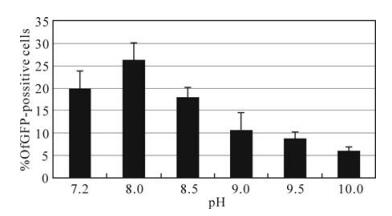
To study the pathogenesis of HearNPV ODVs in vitro, we used HaCXW1, which expresses a GFP marker (3). Purified HaCXW1 ODVs were added into HzAM1 cells to examine their infectivity. Since ODVs are normally released in alkaline conditions in the lepidopteran midgut and established a primary infection in epithelial cells, PBS with different pH values (7.2, 8.0, 8.5, 9.0, 9.5) was used to provide an alkaline environment for ODVs in HzAM1 cells. At 72 hpi, green fluorescence was observed in all cases indicating ODV infectious at all the pH values used in the experiment. However, the infection efficacy expressed as percent of GFP positive cells under different alkaline condition (Fig. 2). At pH 8.0 the infection reached the highest with 26.3% GFP positive HzAM1 cells, while there were less GFP positive cells both at lower and higher pH. These results showed that ODV could infect HzAM1 cells in a pH dependent manner with a pH 8.0 being optimal.
-
ODVs penetrating cultured cell might use the same mechanism as ODVs in the midgut. We examined if the four per os infectivity factors (P74, PIF-1, PIF-2 and PIF-3) are involved in ODV infection in vitro. HaCXW1 ODVs was first neutralized with either αp74, αPIf-1, αHa132 or αPIF-3 and then introduced into HzAM1. Normal rabbit serum was used as control. In a mock-neutralization assay, 19% of the cells were GFP 72 hpi. However, there were 0.70%、0.87%、0.70% and 0.79%, GFP positive cells when ODV were only neutralized by αp74, αPIf-1, αHa132 and αPIF-3, respectively (Fig. 3). Thus, each antiserum neutralized ODV infectivity. However, the infectivity was not significantly different when ODVs were neutralized with any two or three of four antiserums (data no shown). It is note worthy to indicate that when ODVs were neutralized with all four antiserums, the infectivity decreased to 0.08%. In other word, ODV lost its ability to infect.
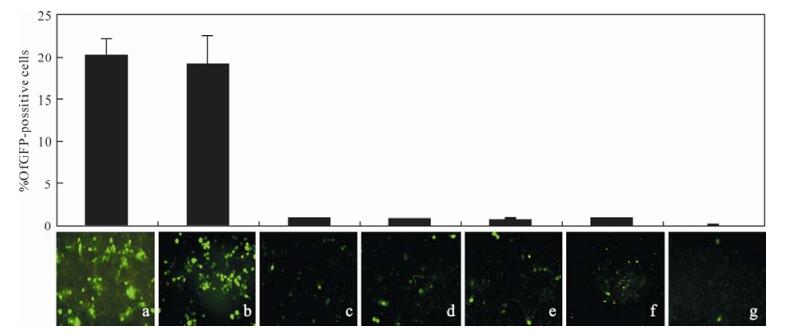
Figure 3. The pictures and percent of GFP positive HzAM1 cells infected with HaCXW1 ODVs, which neutralized by antibodies were examined under fluorescence microscopes at 72 hpi. HzAMI cells were infected with ODVs without (a) or with neutralization of normal serum (b), αP74 (c), αHa111 (d), αHa98 (e), αHa132 (f) or mixture of αHa98、αHa132、αP74 and αHa111 (g).
Expression HearNPV p74, pif-1 and pif-3 and preparation of antiserum
ODVs infect HzAM1cells at different pH values
Virus neutralizations
-
ODVs of baculovirus initiate primary infection in larval midgut epithelial cells, leading to a secondary infection with subsequent BV and OB production. ODV interaction with the microvilli of target cells is of interest not only because of its fundamental importance in baculovirus host range determination, but also because it provides an unusual opportunity to study viral binding and fusion events in a highly alkaline environment. However, due to the lack of an in vitro system, it is difficult to determine the molecular mechanism involved in ODV attachment and penetration of susceptible cells in comparison to BV. Here we demonstrated that HearNPV ODV successfully infected HzAM1 cells in vitro even at low infectivity.
Horton and Burand (9) have reported that ODVs of LdMNPV can infected IPLB-LdEIta cell lines and ODV fusion to cell membrane was found to be greatly enhanced by alkaline conditions, illustrating the specificity of the interaction under alkaline conditions. We found that ODVs of HearNPV also infects HzAM1 cells in vitro. The highest infectivity is at pH 8.0, which is close to the pH value in the microvilli of midgut epithelial cell. However, the infectivity decreased when pH value was more than 8.0. It was reported that the normal pH in the midgut of cotton bollworm is about 10.5, whereas the pH is about 8.5 in the microvilli of midgut epithelial cell (6). So, HearNPV ODV optimally recognizes and binds to special receptors of insect cells in alkaline conditions.
Four proteins have so far been reported to be involve specific receptor on target cells (7), PIF-1 and PIF-2 might function as attachment proteins for ODV binding to primary target cells in the midgut (13). The results presented in this paper demonstrated that P74, PIF-1, PIF-2 and PIF-3 are involved in ODV infection of HzAM1 and might have the same role as in oral infection. Horton and Burand concluded that baculoviral ODV bind to specific receptors in host cells and entry via a nonendocytotic pathway involving direct membrane fusion according to the entry of LdMNPV ODVs into L. dispar cells (IPBL-LdEIta) and in brush-border me-mbrane vesicles (9). The HearNPV ODV-HzAM1 system also provides an efficient tool for further analysis of the molecular mechanisms of ODV infection.













 DownLoad:
DownLoad: