-
Aptamers are high affinity ligands, which can be attained from a randomized pool of molecules by repeated separation and amplification processes to interact with the target, an efficient procedure called "SELEX" (Systematic Evolution of Ligands by Exponential Enrichment) (7, 29). The process through which aptamers have been identified involves repetitively refining the library by partitioning on the basis of selective binding to the target molecule, followed by reamplification. In the past years, several nucleic acid aptamers have been identified (Fig. 1A). Recently, peptide aptamers based on a yeast two-hybrid system were isolated (Fig. 1B) (6). Both the original nucleic acid-based aptamers and the derivative peptide aptamers have the potential to dominantly interfere with specific activities of their target proteins. Thus, aptamers have emerged as valuable new tools to study the biological processes.
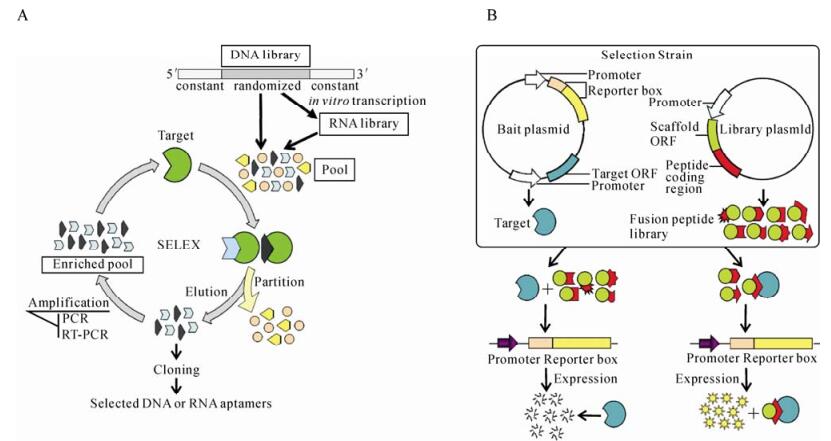
Figure 1. in vitro Selection scheme of aptamers. A: Selection of nucleic acid aptamers. The process involves repeated cycles of binding to the target, partitioning and amplification of 'binders', and commences with a DNA library comprising a randomized sequence flanked by 5′ and 3′ constant regions. For RNA aptamers selection, the DNA library is transcribed in vitro and fold at first, and then incubated with the target protein before the unbound RNAs are partitioned from the bound ones. The latter are eluted, reversetranscribed and then amplified to produce a library of reduced complexity named enriched pool, which is used to initiate the next cycle of the process. After several rounds of selection, the products are cloned once it is judged that the proportion of binding sequences has risen to an appropriate level and are analysed individually. B: Selection of peptide aptamers. The library plasmid expressing fusion peptide is introduced into the selection strain, which contains the compatible bait plasmid that expresses the target protein and the reporter gene, respectively. The expressed activity of the reporter gene loses due to the interaction between the target and the reporter protein, while the activity can be detected or measured in the case that the target is previously bound by the fusion peptide.
Application of traditional drugs usually causes side effects and drug resistance of pathogenic agents. A good example is the emergence of drug-resistant variants of hepatitis B virus (HBV) following lami-vudine treatment. The produced YMDD (tyrosine-methionine-aspartate-aspartate) mutations of the viral reverse transcriptase ultimately render the therapy ineffective. These problems indicate the necessity and urgency of developing new therapeutic strategies that are able to bypass drug resistance. Aptamers can overcome these drawbacks and are now widely accepted as substitutes for traditional compounds.
Additionally, properties such as high affinity and specificity also highlight the huge potential of aptamers for diagnostic and therapeutic applications. Furthermore, aptamers have several advantages over traditional antibodies such as stability, efficiency to attack targets and the benefit of intracellular delivery. Hence, they have been broadly selected to develop antiviral agents for therapeutic applications against hepatitis B and C viruses (HBV, HCV) (2, 3, 12, 14, 15, 18, 28, 30) and other pathogenic agents (5, 9, 10, 23). Another advantage is that prior knowledge about target proteins is not required (11). Thus, aptamers are becoming extremely useful for discovery of new drugs. The first commercialized aptamer drug, Pegaptanib, a targeted anti-VEGF (Vascular endothelial growth factor), has already been applied to the treatment of ocular vascular disease (21).
HTML
-
Hepatitis B virus (HBV), a member of the Hepadnaviridae family, is responsible for infections that cause B-type hepatitis (20). With about 2 billion people having been infected by the virus and 350 million or more being chronic carriers (WHO Fact sheet no. 204), HBV has become one of the most dangerous human viral pathogens. It is a small enveloped virus that replicates by reverse transcription of an RNA intermediate-the pregenomic RNA (pgRNA). Until recently, relatively little was known about the mechanism of initiation in HBV replication due to its several unique features (1). Fortunately, duck HBV (DHBV) provides a valuable model system to study HBV replication (25) because the general features of genome replication are highly conserved.
The epsilon (ε) stem-loop on the pgRNA, consisting of a lower stem, an apical loop, an upper stem and a bulge region, is essential for the encapsidation of the pgRNA and initiation of reverse transcription (12). Both events are mediated by binding of reverse transcriptase (P protein) plus cellular chaperones to ε RNA (Fig. 2A).
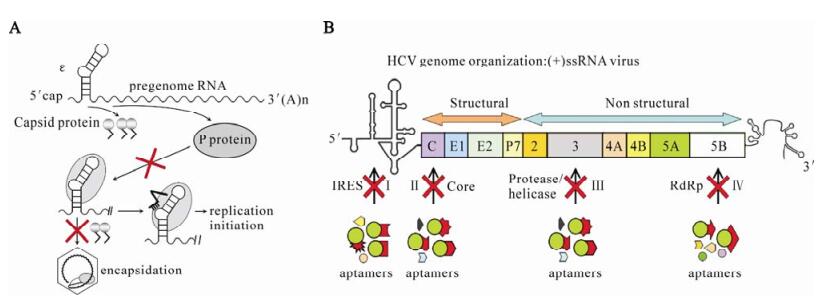
Figure 2. Selection of aptamers against DHBV and HCV. A: The ε stem-loop on the pgRNA is essential for the encapsidation of the pgRNA and initiation of reverse transcription in HBV. Both events are mediated by binding of reverse transcriptase to ε RNA. Hu et al (12) developed an in vitro SELEX to select strong P protein binders from pools of randomized Dε RNAs (A1). Tomai et al (27) developed a system for selecting peptide aptamers that specifically bound to the DHBV core protein and blocks DHBV replication by inhibiting viral capsid formation (A2). B: The HCV-positive strand RNA genome contains a single ORF flanked by 5′-and 3′-untranslated regions and translated as a polyprotein. The polyprotein is processed into at least 10 mature proteins and some of them become valuable targets such as IRES (BⅠ), Core (BⅡ), NS3 (BⅢ) and NS5B (BⅣ).
Recently, we developed an in vitro SELEX to select strong binders from pools of randomized Dε RNAs (12), in which 4 positions proceeding, 4 positions following the apical loop (corresponding sites 2583-2586 and 2594-2597 of the wildtype DHBV genome) were introduced, accounting for nominally 48(65 000) variants. Selection is based on in vitro translation of the His-tagged DHBV P protein, followed by Ni2+ chromatography to isolate the fusion P protein complex together with the bound RNA. Bound RNAs are then eluted, and re-amplified by RT-PCR during which a T7 promoter is reconstituted. The RT-PCR products are used for subsequent rounds of in vitro transcription and selection.
After extensive optimization of the conditions, 9 successive rounds were completed; then individual sequences were isolated by cloning, and separately tested for in vitro activities of P protein binding and priming. Two selected aptamers (S1 and S2) showed 25-40 fold higher affinity to the P protein than the wildtype but only S1 supported protein-priming, and 4 key ε residues (G2586, C2594, U2590 and U2604) have been proved essential for the efficient binding and priming of the P protein. To investigate possible consequences in the context of the viral replication cycle, both S1 and S2 were introduced into complete DHBV genomes pCD16 controlled by the CMV promoter and transfected into LMH cells. Consistent with those data achieved in vitro, S2 did not support DNA synthesis whereas no significant differences were seen between wildtype and S1 in terms of capsid formation and DNA quantity or quality. The result demonstrates that S2 is able to be used as potentially decoy for antiviral therapy. Further experiments in the animal model are under investigation.
Recently, Tomai et al (27) developed a selection system for peptide aptamers that specifically bound to the DHBV core protein, and PA34 showed strong activity of blocking DHBV replication by inhibiting viral capsid formation (Fig. 2A).
-
Hepatitis C virus (HCV), a positive-strand RNA virus of the Flaviviridae family, is a major causative agent of post-transfusion non-A and non-B hepatitis. About 170 million people are estimated to be infected with HCV worldwide (16), and the infection often progresses to chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. The HCV-positive strand RNA genome contains a single open reading frame (ORF) flanked by 5′-and 3′-untranslated regions and is translated as a polyprotein of about 3010 amino acids (24). The polyprotein is processed into at least 10 mature proteins which are believed to be important for propagation of the HCV offspring. Thus, they are valuable targets for development of antiviral therapies (Fig. 2B).
The viral-encoded nonstructural 3 (NS3) protein, a multifunctional enzyme with protease, nucleoside triphosphatase and helicase activities, is essential for viral replication and maturation by cleaving the nonstructural regions of the viral polyprotein. Some laboratories isolated RNA aptamers against NS3 based on an in vitro genetic-selection strategy (Fig. 2B Ⅲ) (1, 8, 17, 31). The RNA aptamers acquired showed the ability to effectively inhibit the proteolytic and helicase activity or the protease activity of NS3. Lately, Umehara et al created advanced dualfunctional (ADD) aptamers that were more effective inhibitors of NS3 protease/helicase activity by attaching the structural domain of the helicase aptamer to the 3′-end of the dual functional aptamer NEO-Ⅲ-14U or the protease aptamer G9-Ⅱ via an oligo(U) (30). With the aim of inhibiting NS3 protease activity, Trahtenherts et al. (28) also used a novel bacterial genetic screen to isolate NS3-inhibiting peptide aptamers. Seven NS3-inhibiting peptide aptamers have been isolated and characterized by investigating the phenotypic changes that SEAP-secreting subgenomic RNA replicons undergo upon intracellular expression of these peptide aptamers, by assaying with real-time RT-PCR and inhibition of SEAP secretion by transfected replicon cells. The peptide aptamers inhibited the NS3 protease activity in vitro and the replication of SEAP-secreting genotype 1b subgenomic RNA replicons upon transfection.
RNA-dependent RNA polymerase (RdRp) is considered as a key enzyme for HCV replication. The nonstructural protein NS5B has an RdRp activity and is the catalytic subunit which ensures the synthesis of the RNA viral genome (19). Thus, NS5B is supposed to be the important target (Fig. 2BⅣ). Based on the genetic selection strategy in vitro, Bellecave et al (2, 3) and Jones et al (14) developed high affinity DNA aptamers against NS5B of HCV and HCV subtype 3a respectively. Biroccio et al also selected RNA aptamers against NS5B for inhibiting polymerase activity (4).
Translation initiation of HCV requires a highly conserved structure known as the internal ribosome entry site (IRES). The IRES consists of four domains (Ⅰ-Ⅳ) and is well conserved among HCV subtypes. The ternary interaction of the IRES, the 40S ribosomal subunit and eIF3 is essential for HCV translation initiation (22, 26). Kikuchi et al. (15) and Konno et al. (16) isolated RNA aptamers capable of binding to the (+) IRES domain Ⅲ and (-) IRES domain Ⅰ respectively. The selected aptamers showed signifi-cant inhibition ability both in vitro and in vivo (Fig. 2BⅠ).
Besides the targets discussed above, RNA aptamers have been selected against the HCV core antigen (Fig. 2BⅡ) (18). Using these aptamers, researchers have developed an aptamer-based biosensor for HCV diagnosis and have detected the core antigen from HCV infected patients' sera with good specificity.
-
Since its discovery in 1990, more and more facts have showed that aptamer is an excellent candidate for diagnostics, bio-analysis, therapeutics and drug discovery. However, the application of aptamers has been forestalled partly by issues about how they can be effectively delivered into target cells (13). Furthermore, several chemical modifications are believed to raise the stability and lifespan of aptamers within the intracellular environment, but accom-panyied with cytotoxicity in many cases. Several reports demonstrated that an aptamer-induced host immune response seems to be weaker than antibody-induced. Obviously, many problems remain to be solved before the commercialization and large-scale application of aptamers.
In summary, aptamers against viral hepatitis have been rapidly developed owing to their advantages compared to antibodies-based therapy as well as development of new efficient selection methods. Presently, the antiviral effectiveness of all selected HBV-and HCV aptamers are being tested in animal models or clinic Ⅰ-Ⅲ trials. In the near future, the successful commercialization of these aptamers will provide novel alternative therapeutic strategies against viral hepatitis.













 DownLoad:
DownLoad: