-
Rabies virus is a nonsegmented, single-stranded negative-sense RNA virus, and belongs to the genus Lyssavirus of the family Rhabdoviridae. The technology for recovering rabies virus from full-length cDNA clone was first developed by Schnell et al (10) and the rabies virus SAD B19 strain was rescued in this study. Since then, RC-HL (6) and HEP-Flury (5) have also been recovered from cDNA clones and the reverse genetics technology has been seen many improvements during this time. Initially, a recombinant vaccinia virus was needed to provide T7 RNA polymerase in the reverse genetic system. Subsequently the system only needed cells that could express T7 RNA poly-merase and did not require the use of vaccinia virus. Ultimately, the system was improved so that T7 RNA polymerase was not needed but instead used RNA polymerase Ⅱ from a variety of cell lines. Therefore, the original low efficiency, cell type restricted technology for the rescue of rabies virus was developed into a high efficiency system and could be applied to a wide variety of cell lines.
The rabies virus genome is approximately 12 kb, comprising five genes that encode the nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and RNA-dependent RNA polymerase (large protein, L) (11). During the course of a rabies virus infection, the viral genome RNA together with the viral RNA polymerase (L and P proteins) and nucleocapsid (N) proteins form the viral ribonucleoprotein (RNP) complex. The RNP complex assembles through the M protein and is surrounded by the external G protein. The N protein is responsible for encapsidation of the genomic and antigenomic RNAs, while the L protein, in cooperation with the P protein, functions as an RNA-dependent RNA polymerase in infected cells. Therefore, in the reverse genetic system of rabies virus, the helper plasmids encoding the N, P, and L proteins must be provided to implement the rescue of the virus (2).
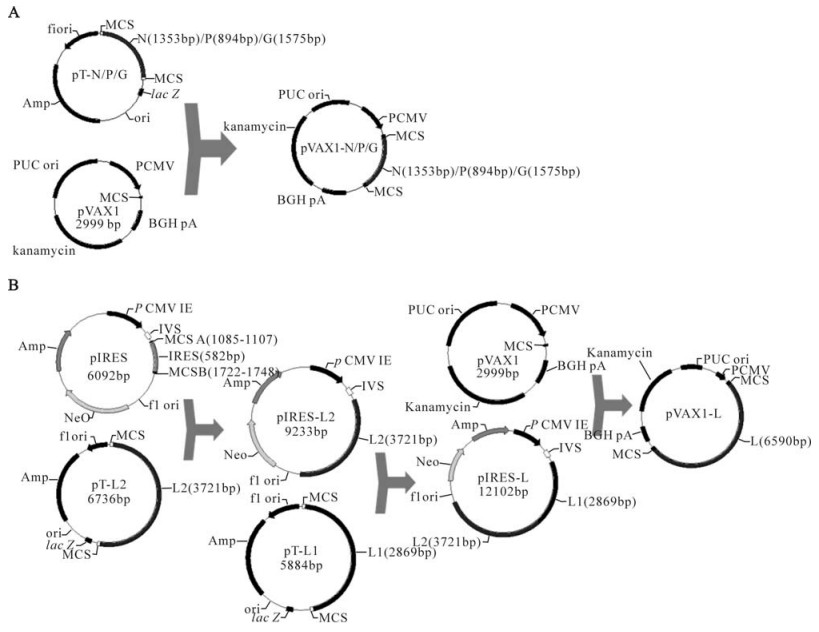
In a previous study, we characterized the complete genome of the rabies street virus HN10 strain (8) and constructed the full-length cDNA clone of this virus (7). In this study, we successfully constructed the helper plasmids encoding cDNAs of the complete open reading frames of the N, P, L, and G genes of rabies street virus strain HN10 which could be directly used in the rescue of this virus.
HTML
-
The rabies street virus HN10 strain was isolated from the brain tissue of an 18-month-old boy with rabies in Yongzhou, Hunan province, China. Mouse neuroblastoma (NA) cells were grown at 37℃ in Dulbecco's minimum essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS).
-
The virus was isolated according to the methods in Ming et al (8).
-
Total RNA was extracted with TRIzol LS Reagent (Invitrogen, USA) according to the manufacturer's instructions. The RNA pellet was resuspended in 70 ml of DNase-, RNase-free sterile water (Promega, USA) and stored at -70℃. For reverse transcription, Ready-To-Go You-Prime First-Strand Beads (Amersham Biosciences, USA) were used to synthesize single-stranded cDNAs. The RT products were then used for PCR. Five fragments were amplified by PCR using the Easy-A High-Fidelity PCR Cloning Enzyme (Stratagene, USA) according to the methods in Ming et al (7) and the primers in Table 1.

Table 1. Primers used in this study
-
All of the PCR products of the expected size were then excised from the agarose gel and purified with the QIAquick Gel Extraction Kit (Qiagen, Germany), following the manufacturer's instructions. The purified PCR products were then ligated with pGEM-T Easy Vector (Promega, USA). The ligation products were used to transform competent Escherichia coli (DH5α or JM109) cells. These cloned products were checked by restriction endonucleoside enzyme digestion and gene sequencing. The five cloned products were named pT-N, pT-P, pT-G, pT-L1, and pT-L2.
-
N, P, and G genes were digested with proper restriction enzymes from pT-N, pT-P, and pT-G, respectively and inserted into the multiple cloning sites in the expression vector of pVAX1. The resulting plasmids were designated as pVAX1-N, pVAX1-P, and pVAX1-G, respectively. The expression plasmid carrying the L gene was constructed with two steps utilizing the unique restriction enzyme recognition site of MluⅠinside the open reading frame of the L gene. Since the restriction enzyme site of MluⅠwas also present in elsewhere in the pVAX1 sequence, the pIRES vector (Clontech, USA) was selected as an intermediate vector to use in the construction of this helper plasmid. The two cDNA fragments of the L gene were cut with the appropriate restriction enzymes from pT-L1 and pT-L2 respectively and inserted into the pIRES vector in turn, and designated pIRES-L. Then the L gene was digested from the pIRES-L plasmid and inserted into the pVAX1 vector. (Fig. 1).
-
For detection of the helper plasmids encoding the N, P, L, and G proteins of rabies street virus strain HN10, restriction enzyme digestion performed by proper enzymes and gene sequencing for open reading frame of N, P, L, and G genes were both used to confirm the correctness of these expression plasmids.
-
To test if the helper plasmids could express the corresponding proteins, pVAX1-N was selected to be transfected into NA cells by using LipofectamineTM 2000 (Invitrogen, USA), according to the manufacturer's instructions. After three day's incubation in 5% CO2, the NA cells were examined for the presence of the N protein by immunofluorescence assay using fluorescein isothiocyanate (FITC)-labeled rabies virus N protein-specific antibody (Millipore, USA).
Virus strain and cells
Virus isolation
Reverse transcription (RT) and PCR
T/A cloning of the PCR products
Construction of the helper plasmids for the reverse genetics system of the HN10 strain
Confirmation of the helper plasmids by restriction enzyme digestion and gene sequencing
Expression of pVAX1-N in NA cells
-
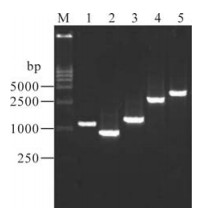
The PCR products were identified on a 1% UltrapureTM Agarose (Invitrogen, USA) gel and visualized by ethidium bromide staining under UV illumination with 15 kb DNA markers (TaKaRa, Japan). There were visible bands at 1 353bp, 894bp, 1 575bp, 2 869bp, and 3 721bp corresponding to the N, P, G, L1, and L2 fragments respectively (Fig. 2).
-
NheⅠ and KpnⅠ were used for restriction enzyme digestion of pT-N, KpnⅠand EcoRⅠfor pT-P, NheⅠand AfiⅡ for pT-G, and PvuⅠfor pT-L1 and pT-L2. The expected fragments were pT-N, 1 353bp and 3 018 bp; pT-P, 894 bp and 3 018bp; pT-G, 1 575 bp and 3018bp; pT-L1, 1 100 bp, 2 000 bp and 2 800 bp; pT-L2, 1 100 bp and 5 680 bp (Fig. 3).
-
NheⅠ and KpnⅠ were used for restriction enzyme digestion of pVAX1-N, KpnⅠ and EcoRⅠ for pVAX1-P, NheⅠ and AfiⅡ for pVAX1-G, MluⅠand NheⅠ for pVAX1-L. The expected fragments were pVAX1-N, 666bp and 3 686bp; pVAX1-P, 666 bp and 3 227bp; pVAX1-G, 666 bp and 3 908bp; pVAX1-L, 666 bp, 2 837 bp and 5 937 bp (Fig. 4).
-
The four plasmids pVAX1-N, pVAX1-P, pVAX1-G, and pVAX1-L were sequenced with the universal primers of T7 and BGH. Comparison of related nucleotide sequences of rabies street virus HN10 strain published in GenBank (accession number: EU643590) with the four plasmids showed that the corresponding gene sequences were all identical. This result showed that the four helper plasmids of pVAX1-N, pVAX1-P, pVAX1-G, and pVAX1-L were successfully constructed.
-
Following three days' incubation in 5% CO2 after transfection, the transfected NA cells were detected by fluorescence antibody of anti-N protein monoclonal antibody (MAb). Fluorescence staining in NA cells showed that the N protein could be expressed in the cells (Fig. 5).
Identification of PCR products of N, P, G, L1, and L2 fragments
Restriction enzyme digestion of pT-N, pT-P, pT-G, pT-L1, and pT-L2
Restriction enzyme digestion of pVAX1-N, pVAX1-P, pVAX1-G, and pVAX1-L
Gene sequencing of pVAX1-N, pVAX1-P, pVAX1-G, and pVAX1-L
Protein expression of the helper plasmid of pVAX1-N
-
Since Schcell et al. (1994) first established a system for rescue of rabies virus from cloned cDNA, three strains of rabies virus, SAD B19, RC-HL, and HEP-Flury, have been recovered from cDNA plasmids (5, 6, 10). However, there are few studies reporting a reverse genetics system for rabies virus from China. One such system was constructed by Ai et al (1) that used the full-length genome cDNA of the HEP-Flurry strain and four helper plasmids with N, P, G, and L provided by Inoue et al (5). Also, Guo et al (3) constructed a full-length plasmid with the G gene at the promoter-proximal position and rescued the new rabies virus by reverse genetics to study the stepwise attenuation of replication and lethality for mice. But the rabies virus used in their study was not isolated from China. In order to better understand the prevalence of domestic rabies and determine appropriate prevention and control measures for this disease, it is necessary to develop reverse genetics technology of rabies virus strains specific to China.
The pVAX1 vector is specifically designed for use in the development of DNA vaccines approved by the FDA. Inoue et al (5) used the pcDNA3.1/Zeo (+) expression vector to rescue the rabies virus HEP-Flury. The pVAX1 vector (4) was generated from the pcDNA3.1 vector after a series of structure optimizations. These optimization features were: (ⅰ) sequences that were unrelated to gene replication and expression of exogenous proteins were removed from the pcDNA3.1 vector so as to reduce the homology of the vector sequence and the human genome sequence, and to minimize the possibility of chromosomal integration; (ⅱ) the ampicillin resistance gene in pcDNA3.1 vector was replaced with the kanamycin resistance gene for selection in E.coli in pVAX1 vector. Since kanamycin is an aminoglycoside antibiotic and is less likely to cause an allergic response in human, the pVAX1 vector is suitable for a variety of mammalian cells; (ⅲ), expression levels of recom-binant protein from pVAX1 are comparable to those achieved with its parent vector, pcDNA3.1. Also, the smaller size of pVAX1 relative to pcDNA3.1 is conducive to transfection into cells.
In this study, we selected the N gene to determine the expression effect because only anti-N protein monoclonal antibody was commercially available and there were not commercial MAbs for the other three proteins. Because sequencing results showed that the nucleotide sequences of rabies street virus HN10 strain published in GenBank (accession number: EU643590) and the corresponding gene sequences of pVAX1-N, pVAX1-P, pVAX1-G, and pVAX1-L were all completely identical we thought that pVAX1-P, pVAX1-G, and pVAX1-L could also be expressed successfully in cells based on the result that pVAX1-N could be expressed in cells.
For initiation of an infectious cycle, full-length positive sense RNA has to be transfected and immediately encapsidated in the transfected cells. Therefore, the viral RNA polymerase (L and P proteins) and nucleocapsid (N) proteins are provided by simultaneously transfecting with plasmids encoding these proteins (known as helper plasmids), resulting in active ribonucleocapsids. Therefore, the rescue of rabies virus needs the participant of the helper plasmids encoding the N, P, and L proteins. Although transfection with pVAX1-G plasmid (encoding viral glycoprotein) is not essential for generation of rescued virus, the supply of G protein occasionally improves yield of the virus (9).













 DownLoad:
DownLoad: