-
Bovine herpesvirus 1 (BoHV-1) is a member of the family Herpesviridae, subfamily Alphaherpesvirinae[2]. It is recognized worldwide as a major pathogen for cattle[29], and is the causal agent of a variety of clinical syndromes with high morbidity and low lethality rates, such as infectious bovine rhinotracheitis (IBR), a severe respiratory tract infection of great economic impact, as well as infectious pustular balanopostitis and infectious pustular vulvovaginitis (IPB/IPV); occasionally, the virus has also been associated with central nervous disorders[1, 23].
BoHV-1 isolates have been classified into subtypes 1 and 2, based on restriction endonuclease analyses (REA) of the viral DNA after virus isolation[7], BoHV-1.1 being mainly associated with IBR and BoHV-1.2 with IPB/IPV[16]. The differentiation between BoHV-1 strains is usually made by the clinico-epidemiological characteristics of outbreaks, usually followed by REA of viral DNA[21, 27] in which REA profiles have led to the subtyping of viruses apparently belonging to the same type, but associated with specific clinical syndromes. Based on genomic and antigenic differences, BoHV-1.2 could also be subclassified as 2a and 2b without strong association with clinical, pathological or epidemiological parameters[4]. In addition, it has been possible to characterize BoHV-1 isolates using monoclonal antibodies (MAbs) against antigenically distinct epitopes identified on the viral glycoproteins (g) gB, gC, gE and gD, some of which are targets for neutralizing antibodies[28]. However further clarification is required to determine which of these could be responsible for the role of immunodominant epitopes in virus characterization[4].
On the other hand, virulence of isolates has long been characterized by one step growth curves, which allow for assessing the growth and replicative capacity of isolates compared to a parental or reference strain, demonstrating the presence of attenuated or highly virulent strains in vitro[3, 20], which has been efficiently corroborated with the pathogenicity of the BoHV-1 in its natural host[12, 13, 17].
In Colombia, BoHV-1 was originally isolated in the 70's; however, there is no clear knowledge about the present BoHV-1 subtypes. The aim of this study was to isolate and characterize field isolates of BoHV-1 obtained from different cattle production systems in the country, to establish which is the acting subtype and to postulate the possible disease mainly associated with it, using also BoHV-1 reference strains to compare the results and correlate REA patterns with monoclonal antibody patterns and in vitro growth kinetics.
HTML
-
The Madin Darby Bovine Kidney (MDBK -ATCC CCL-22) cell line was obtained from the cell repository of the Virology Laboratory of the Microbiology and Epidemiology Group, National University of Colombia; it was maintained at 37℃ in 5% CO2 in minimum essential medium (MEM; Gibco-Invitrogen, Inc. Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Gibco-Invitrogen, Inc. Grand Island, NY) and penicillin-streptomycin-amphotericin (10 000 U/mL; 10 μg/mL; 0.25 μg/mL respectively; Gibco-Invitrogen, Inc. Grand Island, NY). The virus strains IOWA and Colorado-1 (cooper-1 ATCC-V864), harvested from MDBK cells and stored at -70℃ served as reference strains.
-
Five days before taking the samples, seropositive animals were intravenously treated with 200 mg dexametasone (Azium®, Intervet/Schering-Plough Animal Health, USA) to facilitate endogenous virus reactivation[30]. Samples of nasal, ocular, vaginal and preputial swabs were taken using sterile cotton swabs. The swabs were transported chilled to the laboratory in plastic vials with 1 mL of serum-free transport medium. Animal handling and sampling procedures were carried out in compliance with the recommendations of the "Guide for the Care and Use of Laboratory Animals" of the National Research Council (Academy Press, 1996, Washington, USA).
-
Once in the laboratory, the swabs were centrifuged for 20 min at 640 r/min and the supernatants were recovered and filtered through 0.45 μm filters. Subsequently, 200 µL of the filtrate of each swab (two wells per sample) were put in contact with MDBK cell monolayers grown in 24-well plates (USA Scientific, Inc., Ocala, FL) to isolate the virus. After 1 h of incubation at room temperature, 300 µL of MEM, 10% FBS and penicillin-streptomycin-amphotericin were added and the samples were incubated at 37℃ in 5% CO2 for 72 h. The cultures were examined daily under a light microscopy for cytopathic effects (CPE)[18]. Cultures that remained negative for CPE after 72 h were subjected to three additional passages; briefly, cells were detached, freeze/thawed, centrifugated at 1000 r/min and the supernatants were put on a new monolayer with a new incubation period of 72 h[25]. The viruses were harvested from cytopathic positive samples when the CPE reached 70% of the monolayer and were stored at -70℃.
-
The tissue culture infective dose 50 (TCID50) for each BoHV-1 field isolate and reference strain were performed twice in MDBK cells seeded onto 96-well microtiter plates (USA Scientific, Inc., Ocala, FL) at 5 × 104 cells per well. The growth media in the 96-well plates were replaced by 10-fold (10−1-10−10) total virus dilutions made in medium containing 1% FBS with 8 replicas per dilution. After incubation at 37℃ in 5% CO2 for 72 h, the plates were inactivated and fixed with 3.7% formaldehyde in Phosphate buffered saline (PBS) and stained with crystal violet. The infectious dose was calculated following the Sperman-Kaerber method[9].
Plaque assays were used to measure infectious virus particles such as in total virus and in viral growth kinetics. MDBK cells were seeded onto 24-well culture plates (USA Scientific, Inc., Ocala, FL) at a concentration of 2 × 105 cells per well. Serial 10-fold dilutions (10−1-10−7) of viruses, supernatants or cell lysates were performed and added to the cells. After 1 h, the medium was removed and semisolid medium (0.4% agarose prepared in MEM with 2% FBS) was added into each well. Following incubation for 48 h, the plates were inactivated and fixed with 3.7% formaldehyde in PBS and stained with crystal violet. The number of plaques per well was used to calculate the number of plaque forming units (PFU)/mL. Viral plaques were evaluated under a light microscope for each virus isolate.
-
MDBK cells were infected at a multiplicity of infection (MOI) of 10. After 2 h at 4℃, MEM medium was added and the cells were incubated for 30 min at 37℃ to allow virus penetration. The inoculum was then removed, and the cells were washed twice with PBS and incubated with fresh medium. After 1, 3, 6, 9, 12, 15, 18 and 24 h of incubation at 37℃, supernatants containing extra-cellular progeny viruses were collected, clarified by centrifugation (1500 r/min) and frozen. Titrations of these supernatants as measured by the plaque assay on MDBK cells to determine the extra-cellular virus production. Simultaneously, infected cell monolayers were washed twice with PBS in 500 µL of MEM and frozen at -80℃. Intra-cellular virus production was obtained from titrations of the cell lysates. The number of plaques per well was counted, and the PFUs in the original inoculum (PFU/mL) were determined. The results were expressed as the average of at least 2 independent assays with 2 replicates for each assay.
-
For PCR confirmation of the strains, 1 mL of supernatant from infected cultures was collected and the DNA was extracted using an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) mixture, mixed and centrifuged at 12 000 r/min for 5 min. The upper phase (water phase) was transferred to a new tube, and 2.5 volumes of 96% ethanol and 1/10 volume of 3 mol/L sodium acetate pH 5.2 were added. After mixing, the tubes were incubated overnight at –20℃. The DNA was collected after centrifugation at 12 000 r/min at 4℃ for 15 min. The DNA pellets were washed with 70% ethanol, dried, dissolved in 50 µL of ultrapure water and kept at –20℃ before PCR.
For restriction endonuclease assays, viruses were inoculated in T175 cell culture bottles (175 cm2) with MDBK 80% confluent monolayers, at a multiplicity of infection of 0.01–0.1 and incubated at 37℃. When cytopathic effects reached 80–90% (36-48 h), the culture medium was clarified by low speed centrifugation. Viral DNA was extracted from the supernatants using the commercial kit Blood & Cell Culture DNA Midi Kit® (Qiagen®, Hilden, Germany) according to the manufacturer's recommendations. Briefly, the samples were lysed and proteins simultaneously denatured in the appropriate lysis buffer; proteinase K was then added, the mixtures incubated at 56℃, and the lysates were loaded onto a QIAGEN Genomic-tip in which DNA binds to the column. A wash step was performed to remove any remaining contaminant, and pure, high-molecular-weight DNA was eluted, precipitated with isopropanol, dissolved in ultrapure water (Amresco Inc ® Solon, Ohio) and kept at –20℃ before endonucleases treatment.
-
Amplification was performed in a final volume of 25 µL containing: 1 × PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 9.0 and 0.1% Triton X-100), 200 mmol/L of each deoxynucleotide triphosphate (dNTP), 2.5 mmol/L MgCl2, 2.0 mmol/L of each primer and 0.5 U of Taq DNA Polymerase (Fermentas Inc ® Cromwell Park Drive, USA). Forward and reverse primers were respectively: 5'-ATGCAAGGGC CGACATTGGCCGTG-3' and 5'-TTAGGGCGTAGC GGGGGCGGGCG-3', which generates a fragment of approximately 1180 bp. DNA extracted from IOWA-infected cells was used a positive control, and ultrapure water was used as negative control. Amplification was performed in a thermocycler (Bio-Rad ™ Inc Hercules, CA, USA) with the following parameters: initial denaturation at 95℃/10 min followed by 35 cycles of 95℃/30 sec, 60℃/1 min and 72℃/1 min, with a final extension of 72℃/10 min. Finally, 5 µL of PCR product were analyzed by 0.8% agarose gel electrophoresis, ethidium bromide stained (0.5 mmol/L) and visualized by trans-illumination with UV light.
-
Viral DNA was cleaved with Hind Ⅲ under the conditions recommended by the manufacturer (Fermentas®, Vilnius, Lithuania). The digestion products were separated by electrophoresis in 0.6% agarose gels at 30 V using TAE electrophoresis buffer (4 mmol/L Tris-acetate, 1 mmol/L EDTA pH 8.0). The products were visualized using EZ-VISION™ (Amresco® Solon, OH, USA) and analyzed using Gel Doc XR® (Bio-Rad, Hercules CA, USA).
-
To further define antigenic relationships between the bovine herpesvirus isolates, Western blots were performed as reported by Dasika and Letchworth[5]. Briefly, equal amounts of virus-infected cell lysates or mock-infected lysates were boiled and resolved on 8% SDS-PAGE under reducing conditions[14], and the proteins transferred to nitrocellulose membranes (Bio-Rad, Hercules CA, USA). The blots were blocked, incubated 1 h at room temperature with each anti-BoHV-1 mAb (Table 1) according to Marshall et al., [15] (kindly provided by G. J. Letchworth, Madison, Wisconsin) followed by 1 h incubation with secondary monoclonal antibody HRP-conjugated Anti-Mouse IgG. The signals were visualized after treatment of the membrane with SuperSignal West Pico chemiluminescent substrate solution for 5 min (Pierce, Rockford, IL, USA) and exposed to an X-ray film (Kodak®, Rochester, NY, USA) from 0.5-3 min as necessary to detect a signal.

Table 1. Monoclonal antibodies used for Western blot analysis of field BoHV-1 isolates
-
For one step growth curves statistical analysis was performed using the two-tailed paired student's tests to determine significance with 95% confidence intervals. A p value < 0.05 was considered significant.
Cells and viruses
Animal samples
Virus isolation
Virus titration
One step growth kinetics
Extraction of viral DNA
PCR confirmation
Restriction endonuclease analysis
Monoclonal antibody analysis
Statistical analysis
-
From 29 immunosuppressed animals it was possible to isolate 18 virus samples each from different animals (Table 2), most of them (10 samples) were from the respiratory tract and eight were from the reproductive tract. In most cases the CPE was initially visible 12 h post-inoculation, and became characteristic by 36-48 h post-inoculation. In 6 out of 8 samples from a town of the Cauca Valley (designated Cali-3 to 8 in Table 2) it was necessary to perform two passages to observe CPE in MDBK cell cultures; Figure 1 presents the CPE of BoHV-1 field strains on MDBK cells at different times post-inoculation. Supernatants from all the MDBK cell cultures with positive CPE were subject to PCR to confirm the presence of the virus (data not shown).

Table 2. Origin of BoHV-1 isolated viruses
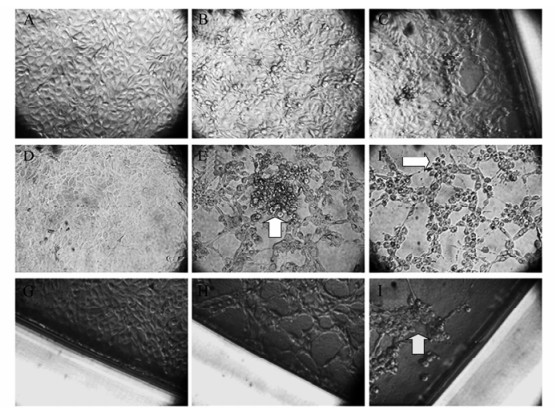
Figure 1. BoHV-1 cytopathic effects at different times of infection in MDBK cell culture. A, D, G: Uninfected controls; B, C: 12 and 24 h.p.i.; E, F: 36 and 48 h.p.i.; H, I: corner of the well at 48 and 60 h.p.i. Original magnification: 25 fold for all samples. Arrows indicate classical CPE, characterized by focal rounding and detachment of cells from the surface of the flask (B), followed by aggregation of the cells into grape-like clusters (E-F, H-I) and by syncytia formation (Ⅰ).
-
Different biological characteristics were evaluated and compared with the IOWA and Colorado-1 reference strains. Plaque sizes were measured in MDBK cells; the plaque size was uniform, between 1 and 2 mm in diameter in almost all field isolates. However, plaque size in the virus isolates designated Cali 1-8 presented a strikingly reduced plaque size, each less than 1 mm in diameter. Both reference strains produced similar plaque sizes to most field isolates (Fig. 2).
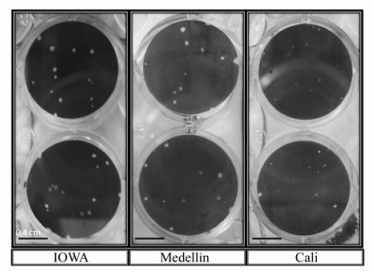
Figure 2. Different BoHV-1 plaque sizes in MDBK cell culture. MDBK cells were seeded on 24-well culture plates at a concentration of 2 × 105 cells per well, infected with each isolate and after 48 h of infection the plates were inactivated, fixed with 3.7% formaldehyde in PBS and stained with crystal violet. A large difference is visible between the IOWA or Medellin plaque sizes and the Cali strains.
The virus titer was evaluated by TCID50 and PFU, in an attempt to highlight differences in virus infectivity compared with the reference strains. The so-called Cordoba and Medellin field isolates had the highest TCID50 titers reaching 1 × 10-8. 25 infectious doses in the Colorado-2 isolate, while the lowest TCID50 titers were as low as 1 × 10-3, 75TCID50 in the Sabana-1 isolate. Highly and mid pathogenic Iowa and Colorado-1 strains had a TCID50 of 1 × 10-7.37 and 1 × 10-5.87respectively.However, whenanalyzingvirus yield by PFU, there was no complete correlation with PFU virus titer. The highest titer was 2.4 × 108 PFU/mL for the Medellin-2 isolate while the lowest was 650 PFU/mL for the Cali-6 isolate. The virus titer for the Iowa and Colorado-1 strains was similar to the Medellin and Cordoba isolates reaching 1.86 × 108 and 2.02 × 107 respectively.
To analyze differences in virus growth, a one step virus curve was performed using a MOI of 10 per MDBK cell, and the virus yields in supernatants or intracellular virus in monolayers were measured. Due to the low PFU virus titer of the Cali and Sabana isolates and the necessity to add 10 viruses per cell, the virus titer was increased by multiple passages in MDBK cells. After 5 cell passages, 2 Cali isolates increased their viral titer to 1.37 × 108 and 5.84 × 107PFU/mL respectively. Likewise, after 4 passages, the Sabana-1 isolate increased its viral titer to 1.12 × 108 PFU/mL. As seen in Figure 3A, the virus yield of the IOWA strain reached the highest titer at 24 h post-infection (6.88 × 109 PFU/mL), as expected due to its highly virulent presentation. Interestingly, some isolates (Cordoba and Medellin isolates) had a highly virulent-like behavior in vitro in comparison to the reference strains, reaching viral titers as high as 6.08 × 108 PFU/mL. Otherwise, the Colorado strain has a medium titer with 1 log less virus yield than the Medellin strain 24 h post-infection. Some strains from Cali had a virus titer as low as 7.42 × 106 PFU/mL (Fig. 3A). In the intracellular virion evaluation, the same difference between native isolates was observed. The Cali isolates showed the lowest intracellular virus titer while the Cordoba and Medellin isolates had the highest virus titers; similar results were found in the IOWA and Colorado reference strains which reached a virus titer of 3.65 × 109 and 9.50 × 108 PFU/mL respectively (Fig. 3B). Other isolates (Sabana-1, Llano-1) had intracellular and extracellular virus productions close to the mean of the assay, showing a virus titer of 107 PFU/mL in both assays (Fig. 3A and 3B).
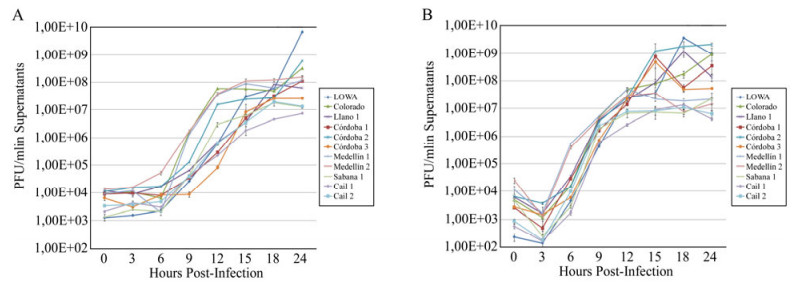
Figure 3. One step growth curve of different BoHV-1 isolates. MDBK cells were infected with different BoHV-1 isolates at a MOI of 10 and the monolayers and supernatants were titrated by the plaque assay at different times after infection to quantify the extracellular virions in supernatants (A) and cell-associated virus on monolayers (B). The values are the means ± SEM of 2 independent experiments with 3 replicates for each experiment.
Also, a shorter eclipse period with faster release of virus in the Cordoba and Medellin isolates was observed in comparison to the Cali isolates (Fig. 3A) with a higher burst size and higher viral load (Fig. 3A and 3B). The Cali isolates were slightly compromised in replication, with the lowest burst size (Fig. 3A and 3B). As expected, there were no difference (p > 0.05) between virus production in BoHV-1 IOWA compared with Medellín and Córdoba strains; however we found significantly lower virus production (p < 0.05) in isolates when comparing IOWA with Cali and Sabana-1 at 18 and 24 hours.
-
The restriction patterns obtained after digestion of each isolate with Hind Ⅲ are shown in Figure 4. The patterns were identical for the reference IOWA and Colorado strains (Fig. 4, lines 5 and 9) in most of the isolates indicating that most of the isolates belong to the BoHV-1.1 subtype. Only the Sabana 1 isolate showed a clearly distinguishable pattern in that one DNA fragment had a slightly lower molecular weight, corresponding to the BoHV-1.2a subtype (Fig. 4, line 13).
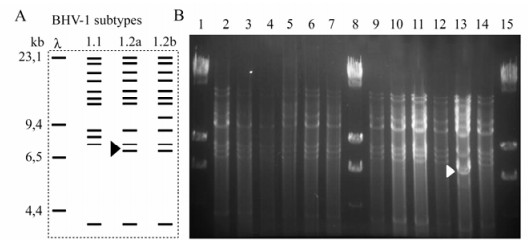
Figure 4. Restriction endonuclease assay of native BoHV-1 isolates. A: Schematic representation of BoHV-1 DNA digested with Hind Ⅲ restriction enzyme showing the pattern of each subtype. B: Restriction endonuclease digestion patterns of BoHV-1 isolates. B. Representative panel of native isolations: Lanes 1, 8 and 15 Hind Ⅲ-digested Lambda Phage; lane 2: Cali 1; lane 3: Cali 21; lane 4: Cali 22; lane 5: Colorado; lane: Cordoba 1; lane 7: Llanos; lane 9: IOWA; lane 10: Med 9; lane 11: Med 12; lane 12: Cordoba 2; lane 13: Sabana; lane 14: Cordoba 3. Arrow-head indicates the difference in BoHV subtype, showing a BoHV-1.2a pattern in the Sabana-1 isolate.
-
To assess the presence of different antigenic viral glycoproteins relevant to natural immune response, Western blots were performed using primary antibodies against the BoHV-1 glycoproteins B, C, D and E. These antibodies were developed by Marshall et al.[15], and are directed against main antigenic areas relevant to natural infection. As observed in Fig. 5, all BoHV-1 isolates presented the same expression for the different glycoproteins in comparison with the IOWA and Colorado reference strains.
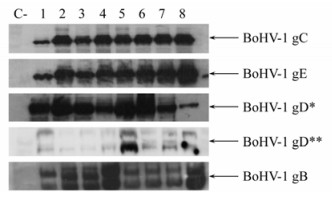
Figure 5. Western blot mapping of different BoHV-1 glycoproteins in native BoHV-1 isolates. Cultures of MDBK cells were infected with different BoHV-1 isolates (MOI = 10). At 24 hours after infection, the cell lysates were prepared as described in Materials and Methods. Anti-BoHV-1 monoclonal antibodies (Table 1) were used according to Marshall et al. [15]. * Epitope group I; ** Epitope group V. Lane 1: BoHV-1 IOWA; lane 2: Cali 1; lane 3: Cali 22; lane 4: Córdoba 1; lane 5: Córdoba 2; lane 6: Medellin 1; lane 7: Medellin 9; lane 8: Sabana; C-Negative control. The image is representative of two experiments for each BoHV-1 isolate.
Animal samples and virus isolation
Biological characterization
Restriction endonuclease analysis
Immunodominant epitope evaluation
-
Many attempts have been made to differentiate strains of BoHV-1, especially those of respiratory and genital origins. Most attempts revealed that different isolates are quite similar with respect to their biophysical and antigenic properties[4, 19]. In Colombia, considerable numbers of cattle farms have shown serological evidence of BoHV-1 infections; using the viral neutralization test it was shown that more that 75% of the sampled population of cattle herds have neutralizing antibodies against BoHV-1 infection[25]. However, there is as yet no knowledge about the present BoHV-1 subtype and the characteristics of the virus present in cattle herds.
From the present results described here, one of the most important findings is that there is a big difference in the in vitro growth behavior of the isolates in MDBK cells. By PFU and TCID50%, the Cordoba and Medellin isolates exhibit a highly virulent-like behavior with virus titers similar to those reached by the IOWA strain (Table 3), which is a highly virulent strain based on its in vitro and in vivo replication behavior[11, 17]. On the other hand, the Cali isolates had the lowest virus titers, as low as 650 PFU/mL; some of these required a specific cell culture adaptation process to increase their viral titer to perform the one step grow curve at a MOI of 10. Although there were no differences in CPE between the strains, in agreement with differences in viral titers in vitro, the plaque size was uniform and bigger for almost all the isolates while the Cali (1-8) isolates had a smaller plaque size (Fig. 2). This result could indicate that these strains are naturally attenuated strains in comparison with other BoHV-1 isolates, as reported for other herpesviruses that infect bovines and equines[6, 8, 10], or it could indicate that those isolates behave in vitro in an attenuated-like manner.

Table 3. Virus titers determined by TCID50 and PFUs for different BoHV-1 isolates and for the reference IOWA and Colorado strains.
The one step viral growth curve also shows that the Medellín and Córdoba isolates have a highly virulent-like behavior, while in the same experiment Cali isolates presented diminished replication with the lowest burst size compared to other isolates and reference strains, again indicating a possible attenuated-like behavior[6, 8].
However, to confirm that some of the isolates are highly virulent and others are possibly attenuated, it is necessary to perform in vivo analyses as reported by Smith et al. for the highly virulent V592 strain of Equine Herpesvirus-1[26], or using the classical rabbit or bovine models of pathogenicity of infection as shown previously for BoHV-1[11].
No differences were found among the isolates of the present study (Fig. 5) in the presence of the main immunodominant viral epitopes of glycoproteins, confirming the high degree of homology between these isolates and indicating that there were no differences in antigenicity (Fig. 5). As reported previously[4], MAb analyses are not really effective for establishing BoHV-1 differences; however as opposed to other studies, in which it was not possible to determine which glycoproteins of the virus were evaluated, in this study a panel of MAbs directed against the main inmunodominant epitopes was used. The study of different epitopes is important to evaluate and predict the neutralizing response to those strains in vivo and in some cases to differentiate BoHV-1 from BoHV-5, which belongs to the same viral subfamily and was previously grouped as a subtype of the BoHV-1[22].Marshall et al[15], developed these antibodies against different epitopes important for natural infection; however, 3 of 5 BoHV-1 gD antibodies could not be evaluated because they are conformational epitopes[5] and Western blots in denaturing conditions were used here.
The restriction pattern found in the native isolates were similar to those described in previous reports[4, 30], and as reported by other authors[24] who observed no direct relationship between clinical patterns and restriction endonuclease profiles. In the present study, BoHV-1.1 was isolated from the respiratory or reproductive tracts of bulls as of cows, confirming that any virus subtype can be spread equally from males to females or vice versa and that there is no relationship between the BoHV-1 subtype and clinical presentation.
It is very important to know that the BoHV-1 isolates evaluated in this study may be representive for the more prevalent areas in the country and permit predictions that some isolates have a highly virulent-like behavior, representing a high risk to the bovine production in the country.
Taking into account the fact that no monovalent BoHV-1 vaccines exist in Colombia, the pool of our results permit us to initiate a new area of research to try and develop new inactivated vaccines. These could be manufactured with native strains as seed using strains that grow efficiently in cell culture without the necessity of spent time on the adaptation to the laboratory[11] or on the other hand, using the strains that have an attenuated-like behavior as live attenuated vaccines.













 DownLoad:
DownLoad: