-
Influenza A Virus (Ⅳ) belongs to the family Orthomyxoviridae[9]. Other members of the family include influenza viruses type B and C, which infect only humans. The virus has an envelope with a host-derived lipid bilayer and is covered with about 500 projecting glycoprotein spikes with hemagglutinin (HA) and neuraminidase (NA) activities. There are 16 subtypes of HA and 9 subtypes of NA.
Avian influenza is a significant threat to the poultry industry. Highly pathogenic AIVs(HPAI) belonging to H5 and H7 subtypes have caused respiratory disease with nearly 100% mortality in poultry[16]. Accidental genetic reassortments of RNA segments from different subtypes of avian, swine and human IVs are capable of inducing a global pandemic[16]. An HPAI H5N1 was reported to be transmitted from birds to pigs and humans leading to significant mortality[3]. The virus was spread by wild free flying birds which increase the threat of a pandemic.
The prerequisite for controlling the disease is rapid and accurate identification of this virus and subtyping. The method recommended for definitive antigenic subtyping of influenza A viruses by the World Health Organization (WHO) Expert Committee involves the use of highly specific antiserum, prepared in an animal giving minimum nonspecific reactions, directed against the H and N subtypes. The agar gel immunodiffusion (AGID) test and enzyme-linked immunosorbent assays (ELISAs) remain the diagnostic assay most utilized for detection of AIV antibodies in commercial poultry worldwide and are considered the " standard" by the World Organization for Animal Health[11]. AGID test detects antibodies to two influenza virus proteins, NP and M1, which are highly conserved and type specific. There is a sensitive and specific ELISA that demonstrates nucleoprotein of type A influenza virus using a monoclonal antibody against type A influenza nucleoprotein. Although these assays detect antibodies against all AIV subtypes, it does not distinguish subtypes amongst them. Reverse transcriptase polymerase chain reaction (RT-PCR) and real time RT-PCR (rRT-PCR) have been developed as rapid detection tests for AIVs[2, 7, 8, 13]. However, the single RT-PCR only recognizes one specific subtype of AIV gene. Multiplex RT-PCR could only detected limited subtypes of AIV [12]. Studies have demonstrated that using multiplex assay for typing and subtyping of Ⅳs[9]However, the numbers of target genes are limited and no more than five. To distinguish the most common subtypes in animals, A GM RT-PCR integrated with luminex technology (GMPLex) was developed for simultaneously differentiating of 7 subtypes of avian influenza A viruses.
HTML
-
Inactivated Avian Influenza A virus/Chicken/HK/ HI/1997(H5N1), A/Chicken/ HK /921/1997(H9N2), A/PFV/Restock/1/1934(H7N1) A/PFV/Restock/1/1934(H7N1), A/Swine/SZ/111/2005(H1N1), A/Swine/SZ /321/2005(H3N2) were obtained from Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Qiagen one-step RNA PCR kit, DNA fragment purification kit, QIAamp viral RNA Mini-prep kit, Qiagen one-step RT-PCR kit and EZ1 virus mini kit2.0 were used. Liquichip carboxy bead from Luminex(Valencia, CA), Luminex calibration bead mix and Luminex control bead kit from Qiagen (Valencia, CA).
-
To design a set of primers and probes specific to each single HA and NA subtype, we compared nucleotide sequences of all subtypes of HA genes retrieved from the Genbank. Sequences from regions that were conserved in only a single subtype were chosen for the primer design. A total of 9 sets of primers and probes were designed by this strategy for GM RT-PCR amplification (Table 1). Briefly for H5, for example, probes were selected from highly conserved regions of target genes specific for HA gene of H5 influenza viruses, by the sequence data available in the Genbank. The sequence data were generated using the sequence analysis of the influenza database at http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi?go=1. AIV HA gene of H5 subtype from different years was selected, but mainly after 2004. As well as different countries mainly from Asia, different hosts but mainly avian, especially chicken and duck, Nine hundred twenty six strains were selected and analyzed, and made multiple alignment with software DNAStar 5.06(DNASTAR, Inc., Madison, WI). It selected 20 most conservative oligomucleotides in the HA gene of H5 subtype as detection probe for Luminex (Fig. 1). The Tm of probe was kept around 56~58 ℃ and labeled 5' with –NH2.

Table 1. Primers and Probes of GMPLex for each subtype of influenza virus
We designed nested degenerated primers separately before and after probes. There was a pair of universal primer tagged designed primers, and the 5' of downstream reverse primer was labeled with biotin. To amplify HA gene of all the H5 strain, the 3' of each primer should have at least 5 conservative bases. All the above primers and probes had been tested by a software PrimerExpress3.0 (Applied Biosystems Inc. Foster City, CA).
Using the same procedure described above, we designed corresponding nested-degenerated primer and probe targeted highly conserved fragments for NS gene and M gene against all subtype Ⅳ, HA gene against H1, H3, H7 and H9 subtype, and NA gene against N1 and N2 subtype (Table 1) manually.
-
Viral RNA was extracted from the supernatant of the allantoic fluids from different birds by using QIAamp RNA extraction kit (Valencia, CA) according to the manufacturer's instruction. The RNA was eluted from the QIAspin columns in a final volume of 50 μL of elution buffer and immol/Lediately stored at -70 ℃ until used.
-
The General multiplex RT-PCR (GM RT-PCR) was carried out in a reation mixture (50 μL volume) containing 1 × RT-PCR reaction buffer (Qiagen), 0.4 mmol/L each of four dNTPs (Qiagen), 2.5 mmol/L MgCl2(Qiagen), 2 μL one step RT-PCR enzyme mix (Qiagen), 0.6~2.4 μmol/L of super primer mixture, 0.05~0.2 μmol/L single primer, 10 U RNase inhibitor (Qiagen), 2 μL of RNA template. The GM RT-PCR conditions were step 1, reverse-transcription, 50 ℃ for 35 min; step 2, Inactivate reverse-transcriptase and hot-start, 95 ℃ for 15 min; step 3, enrich, fifteen cycles of 94 ℃ 30 s, 52 ℃ 1 min and 30 s, and 72 ℃ for 1 min; step 4, adding tail, 6 cycles of 94 ℃ 30 s, 70 ℃ 1 min and 30 s; step 5, amplification, 35 cycles of 94 ℃ 30 s, 55 ℃ 30 s, and 72 ℃ for 30 s, followed by a final 72 ℃ for 10 min were done. The products were subjected to electrophoresis on a 1.5% agarose gel and visualized under UV.
-
2 μL of viral RNA from each strain, with any two combination mixtures from the following strains, A/Chicken/HK/HI/1997(H5N1), A/Chicken/HK/921/1997(H9N2), A/PFV/Restock/1/1934(H7N1), A/Swine/ SZ/111/2005(H1N1), A/Swine/SZ/321/2005(H3N2), GM RT-PCR was then done by the protocol described in 2.2.4.
-
Viral RNA of 2μL from each of the A/Chicken /HK/HI/1997(H5N1), A/Chicken/HK/921/1997(H9N2), A/PFV/Restock/1/1934(H7N1), with combination mixtures, then the GM RT-PCR followed the protocol described in 2.2.4.
-
Viral RNA of 2μL from each of the above 5 strains, with combination mixture, set the total volume as 50 μL and then GM RT-PCR followed the protocol described in 2.2.4.
-
According to the sequence of fluorescent coding microspheres suggested by manufacture, 10 of them were selected, which had the lowest fluorescent signal misreading probability, binding with the following probe according to table 2, NS gene, M gene of Ⅳ, HA gene of AIV subtype H1, H3, H5, H7 and H9, NA gene of N1 and N2. The couple reaction of fluorescent code microsphere to probes of AIV was done according to manufacturer's instruction. Briefly, all reagents were recovered to room temperature (RT), probes diluted to 0.2 mmol/L with 0.1 mol/L MES solution (pH 4.5). All the following operations were done in a dark room. The fluorescent coding microspheres storage solution were votexed evenly, then 200 μL solution (5×106) was aspirated to 1.5 mL eppendorf tube, centrifuged at 10 000 r/min for 5 min, supernatant discarded, added to 50 μL 0.1 mol/L MES solution (pH4.5), vortexed and added to the appropriate amount of 0.2 nmol probes. The solution was mixed and 10 μL of fresh made EDC solution (10 mg/mL) was added and mixed and incubated for 30 min. This step was repeated, 1 mL Tween -20(0.02% w/v) was added to wash the microsphere, centrifuged at 10 000 r/min for 10 min, supernatant discarded, 1.0 mL SDS(0.1% w/v) added, centrifuged at 1 000 r/min for another 10 min to precipitate the microsphere. The appropriate amount of 0.1 mol/L MES (pH 4.5) was added. The microsphere was counted by using blood cell counting assay, the concentration of the microsphere calculated and then diluted the microsphere concentration to 5 000 microspheres per 1 μL with MES solution, and this solution stored at 2~8 ℃.

Table 2. Microspheres Code and its corresponding AIV probes
-
To confirm whether the various IVs subtype probes coupled microsphere could hybridize with the corresponding product of GM RT-PCR, GM RT-PCR products were taken from IVs subtypes of A/Chicken/ HK/HI/1997(H5N1), A/Chicken/HK/921/ 1997(H9N2), A/PFV/Restock/1/1934(H7N1), A/Swine/ SZ/111/2005 (H1N1), A/Swine/SZ/321/2005(H3N2), hybridized with the corresponding probe coupled microspheres according to the following protocol. All reagents were recovered to room temperature (RT) and all the following operations were done in a dark room. First 50 μL of fresh made GM RT-PCR product was purified by using Qiagen DNA Fragment Purification Kit (Valencia, CA), diluted the purified PCR product to 36 μL. the probe coupled microspheres prepared in step 2.3.1 were taken with 10 μL respectively and further diluted with 1.5×TMAC hybridization solution to 200 microspheres per 1 μL. then probe coupled microspheres working solution were added to the 96 U microplate and set 17 μL of TE solution (pH 8.0) as blank control. purified and diluted GM RT-PCR product with 5 μL were added to the wells, the microplate coated with the film and the reaction in a PCR apparatus to hybridize was performed. The reaction condition was set as 95 ℃5min, 52 ℃ 15 min, then centrifuged at 10 000 r/min for 3 min to collect the microspheres. Fresh Reporter Mix was done by diluting streptavidin-R-phycoerythrin to 10 μg/mL in 1×TMAC Hybridization Solution. Reporter Mix of 25 μL was added to each well and gently mix by pipetting several times. The reaction plate was incubated at the hybridization temperature for 5 min. Total 50 μL at hybridization temperature were done on the Luminex analyzer according to the system manual for analysis.
According to the determination criteria recommended by Luminex, if the microspheres of each fluorescent is no less than 20 and the fluorescent number of the blank control is not more than 3 000, the results can be standardized. Luminex qualitative ratio result (LQRR) equal to Median florescence intensity (MFI) divided by MFI of blank control, that is LQRR=MFIS/MFIB. The LQRR≥3, is positive; if the LQRR is between 2 and 3, is suspicious; and LQRR≤2, is negative.
-
Ten fluorescence coded microsphere with 100 μL of respectively were diluted with 1.5 TMAC hybridization solutions to 200 microspheres per 1 μL. It was vortexed and made into a mixture and other steps were the same as the protocols described in 2.3.2. The set machine could detect 10 fluorescence coded microspheres simultaneously, determination criteria was the same as the methods described in 2.3.2. In order to verify whether GMPLex detection results were identical with subtypes of different AIV, the above 5 different AIV GM RT-PCR products were taken respectively, then 2 or 3 combinations of the GM RT-PCR products were used, and 5 of them were hybridized with the 10 fluorescence coded microspheres mixtures, following the conditions described in 2.3.2.
-
The preparation of RNA transcripts of different subtypes of Ⅳ for GMPLex was conducted with the stain of A/Chicken/HK/HI/1997(H5N1). The sensitivity of the GMPLex was evaluated with different (50% embryo lethal dose, ELD50) ranging from 1 to 109 with ten-fold dilutions[14]. The assay of each Ⅳ dilution was conducted in triplicates. The specificity of the GMPLex was examined by using RNA extracted from 83 viral samples delivered from Hongkong; all of the samples were investigated with virus isolation and the GMPlex respectively.
Virus strain and major reagents
Design of the probe and primer
RNA isolation
Development of a GM RT-PCR for the detection of single subtype of Ⅳ
Development of a GM RT-PCR for the detection of multiple subtypes of AIV
Development of a GM RT-PCR for the detection of two subtype IVs
Development of a GMT RT-PCR for the detection of H5, H7 and H9 subtype
Development of a GMT RT-PCR for the detection of five subtypes of Ⅳ
Development of a GMPLex to detect AIV
Binding the oligonucleotide probe to Luminex carboxylated fluorescent coding microspheres by one-step EDC coupling reaction
Development of a GMPLex to detect single subtype of Ⅳ
Development of a GMPLex to detect multiple subtypes of Ⅳ
Sensitivity and specificity of GMPLex high throughput test
-
The nucleotide sequences of NP gene and M gene were highly conserved in all subtypes of influenza viruses. Five virus strains were all amplified with 174 bp fragment targeted M gene and 145 bp fragment targeted NS1 (data not shown). For a single subtype, only one of the GM RT-PCR reactions are the product of expected size. The size of PCR product ranged from 126 bp to 245 bp, depending on the HA and NA subtype. The specificity of GM RT-PCR indicated it was positive for H5 gene and N1 gene of A/Chicken/ HK/HI/1997(H5N1), H1 gene and N1 gene of A/Swine/SZ/111/2005(H1N1), H3 gene, N2 gene of A/Swine/SZ/321/2005(H3N2), H7 gene of A/PFV/ Restock/1/1934(H7N1), H9 gene of A/Chicken/HK/ 921/1997(H9N2)(Fig. 2, Table 1).
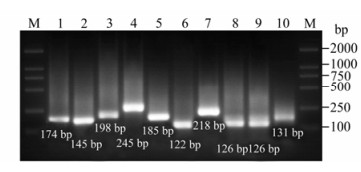
Figure 2. Gel electrophoresis result of GM RT-PCR for single subtype of influenza virus. M: Marker DL 2000; 1: M gene of A/Chicken/Hongkong/SZ-HI/1997(H5N1); 2: NS gene of A/Chicken/Hongkong/SZ-HI/1997(H5N1), 3: HA gene of H1 subtype(A/Swine/Shenzhen/111/2005(H1N1)); 4: HA gene of H3 subtype(A/Swine/Shenzhen/321/2005(H3N2)); 5: HA gene of H5(A/Chicken/Hongkong/SZ-HI/1997(H5N1)); 6: HA gene of H7(A/PFV/Restock/1/1934(H7N1)); 7: HA gene ofH9(A/Chicken/Shenzhen/921/1997 (H9N2)); NA gene ofN1(A/Chicken/Hongkong/SZ-HI/1997 (H5N1)); NA gene ofN1(A/Swine/Shenzhen/111/2005 (H1N1)); NA gene ofN2(A/Swine/Shenzhen/321/2005(H3N2)).
-
GM RT-PCR could amplify expected products from mixture of multiplex Influenza A virus, as A/Chicken/HK/HI/1997(H5N1), A/Chicken/HK/921/ 1997(H9N2), A/PFV/Restock/1/1934(H7N1), A/Swine/ SZ/111/2005(H1N1), A/Swine/SZ /321/2005(H3N2)) (Table 3). The result of amplification was a smear because of strategy of asymmol/Letrical RT-PCR for 9 genes (Fig. 3).

Table 3. Result of GM RT-PCR for multiplex strains mixture of influenza virus
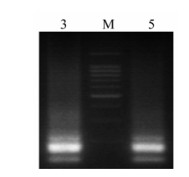
Figure 3. Gel electrophoresis of GM RT-PCR for Multiplex strains mixture of influenza virus. M: 100 bp DNA Ladder Marker (Qiagen, Valencia, CA); Lane 3: A/Chicken/Hongkong/ SZ-HI/1997(H5N1) + A/Chicken/Shenzhen/921/1997(H9N2) + A/PFV/Restock/1/1934(H7N1);Lane 5: A/Chicken/Hongkong/ SZ-HI/1997(H5N1) + A/Chicken/Shenzhen/921/1997(H9N2) + A/PFV/Restock/1/1934(H7N1) + A/Swine/Shenzhen/111/2005 (H1N1) +A/Swine/Shenzhen/321/2005(H3N2).
-
MFI of GM RT-PCR product ranged from 81~18826, according the guideline of Luminex, the LQRR result of H72 was 1 and the result was negative, indicating that there was no specific hybridization between microsphere #38 and H7x GM RT-PCR products. Other probes were positive, the value of LQRR was between 3~27(Table 4).

Table 4. MFI value of GMPLex for single subtype
-
Using mixture of GM RT-PCR product of either 3 mixtures of different subtype strains or 5 mixtures of subtypes, the probe mixture was hybridized. MFI of the product was between 6848~16400 and the MFI of blank control was lower than 3 000. Probe mixture had specific reaction with GM RT-PCR product and LQRR was 3~99.9. (Table 5)

Table 5. MFI value of GMPLex for multiplex strain mixture
-
The sensitivity of the GMPlex was determined by testing a serial dilution of allantoic fluid of the A/Chicken/HK/HI/1997 (H5N1). GMPLex could detect H5 fragment or N1 fragment amplified by single pairs of primers GM RT-PCR in 10-5(equivalent to 280 ELD50), which was tenfold less compared with the sensitivity of real time RT-PCR of the national standard in 10-5(equivalent to 28 ELD50). H5 and N1 were subtype together in 2800 ELD50 (Table 6). When Avian Influenza virus A/Chicken/HK/HI/1997(H5N1), A/Chicken/HK/921/1997(H9N2), A/PFV/Restock/1/1934 (H7N1) mixed together, its detection sensitivity was 2800 ELD50, 28000 ELD50 and 280 ELD50 respectively.

Table 6. Comparison of GMPLex for single type and single strain
-
The specificity of GMPLex assay was evaluated with the main infectious respiratory diseases including Newcastle diseases virus(NDV) (LASOTA), infectious bronchitis virus(IBV) (4/91), infectious laryngotracheitis virus(ILTV) (isolation strain), infectious bursal disease virus(IBDV)(GX strain), marker's disease virus(MDV) (WH strain) and main bacteria infectious diseases including E.coli O157, fowl cholera and Salmonella pullorum. There was no cross reactions, the MFI was lower than 3000 and the LQRR value was lower than 0.88 (below the 1). Therefore, the assay constructed was specific.
-
To evaluate the clinical sensitivity of the GMPLex method, a total of 83 samples derived from vaccines, detection antigens and separations, which were sequenced, were re-tested by GMPLex. The results indicated that the sensitivity and specificity were identical with virus isolation (Table 7).

Table 7. Comparison of virus isolation and GMPLex for detection of Ⅳ in 83 clinical samples
GM RT-PCR for single subtype of influenza virus
GM RT-PCR for multiple strains mixture of Ⅳ
GMPLex for single subtype of influenza virus
GMPLex for multiple strains mixture of influenza virus
Sensitivity of the GMPLex
Specificity of the GMPLex
Evaluation of the GMPLex assay using clinical samples
-
Avian influenza (AI) is a highly contagious disease in poultry and outbreaks can have dramatic economic and health implications. The rapid spread of highly pathogenic H5N1 AIV throughout Asia, Europe, and Africa and its zoonotic potential pose for both public health and the economic integrity of the poultry industry[1, 4]. Thus, there is a critical need for comprehensive and sensitive assays for AIV diagnosis in poultry, especially to distinguish the outbreak subtypes. More importantly, the unique characteristics of distinct AIV subtypes complicate widespread vaccination and prevention strategies, further increasing the importance of effective surveillance.
It is not only needed to detect its IVs positive, but also it must subtype the isolates, so as to adopt corresponding measures to control the disease. Current subtyping needs to detect virus with each subtype anti-serum separately. There were reports that some groups adopted multiplex RT-PCR to identify subtype simultaneously, but due to the technical shortcoming of multiplex RT-PCR, the number it could detect was limited[12]. This study established a completely new General Multiplex RT-PCR (GM RT-PCR) test method, resolving the chip testing "bottle neck" of having to perform many individual PCR amplifications. The influenza virus subtyping GM RT-PCR test and the LiquiChip test methods were combined to give an Ⅳ GMPLex subtyping test method. The GMPlex subtyping test can satisfy the needs of influenza rapid high throughput testing at ports of entry and exit, but also has erected a platform for more novel Ⅳ rapid throughput tests. We constructed a platform of molecular differential diagnostic (MDD) assay that could identify, differentiate, and pinpoint the offending pathogen associated with a clinical syndrome. Compared with the routine diagnostic methods of RT-PCR or rRT-PCR, the GMPLex assay had more high throughput and better improved platform.
The application of a multiplex RT-PCR assay (an RT-PCR assay for the simultaneous detection of different viruses in a sample), offers a significant time and cost-saving advantage, especially when large numbers of samples are analyzed [12]. The principal challenge in developing a multiplex PCR system is choosing the right primers to overcome primer– dimmer formation. Therefore, the oligonucleotide primers selected for the amplification of Ⅳ nucleic acids were analyzed to ensure that they not only meet the essential criteria for optimal PCR primers [5], but also could be used together in a multiplex reaction under identical amplification conditions. The primers were designed with regard to similar length (20-or 21-mers), similar GC content (Table 1), similar annealing temperature, and as little primer–dimmer formation as possible between all six primers. An annealing temperature of 55 ℃ was evaluated to give maximum product yields and specificity. In addition, "hot-start" PCR conditions were used to minimize primer–dimmer formation. Another prerequisite for a multiplex PCR assay is that the PCR products must have different sizes to be clearly identified and differentiated after gel electrophoresis. The 10 pair of primers when used together in the multiplex reaction, amplified only specific products of the expected sizes ranging from 122~198 bp, respectively(Table 1), which could easily be distinguished by agars gel electrophoresis (Fig. 2).
To achieve rapid subtype identification of influenza virus, universal primers were added to the novel GM RT-PCR test so that one PCR would be able to simultaneously amplify many target fragments and, combined with the luminex high throughput test, an influenza GMPLex rapid and high throughput subtype identification test was developed, which attained the target of rapid, accurate and Ⅳ subtype identification. General Multiplex RT-PCR integrated with Luminex (GMPLex) aimed at the establishment of a rapid method for the screening or detection of not only one subtype of AIV, but the relatively multiple subtypes of AIV in all type A virus, as well as successful detection determined on the primer used for GM RT-PCR discussed above, also depended on the probes used in liquid chip test. The probes were designed with regard to similar length (18-or 25-nt), Tm was 5℃ higher than primer's in order to assure hybridization prior to the primers. The LiquiChip system is a bead-based platform that offers the potential to rapidly assay up to 100 different analytes in a single sample; LiquiChip assays are based on xMAP technology and involve the interaction of immobilized, bead-bound capture molecules with a reaction partner (analyte) in solution. A reporter molecule, specific for the analyte, is used to quantify the interaction[6, 10, 15]. Validation of the specificity of the GMPLex revealed there was no cross-reaction with other avian viruses (data not shown), including NDV, IBV, IBDV, Duck hepatitis virus (DHV), avian entero virus (AEV) and host-derived RNA.
The GMPLex assay specifically detected the targeted viruses, as it was demonstrated by sequencing of the amplification products. Because the assay detected 280 ELD50~2800 ELD50, theoretically it could identify even one single infected chicken within a pool of several hundreds of chicken swab samples. The results of this study indicate that the GMPLex described in this paper is a specific, sensitive and reliable tool for the simultaneous diagnosis of multiple subtypes of AIV, as it was confirmed by virus isolation. Because sensitivity and specificity of the system was very similar to that of the monospecific assays, the GMPLex is a cost-saving alternative to single PCRs in routine diagnostic submissions or surveys.













 DownLoad:
DownLoad: