-
The Human Immunodeficiency Virus type 1 (HIV-1) protein U (VpU) is a membrane-associated "auxiliary" protein, approximately 16 kDa, that is unique to HIV-1 and a subset of the related Simian Immunodeficiency Virus (SIV) [3]. This oligomeric integral membrane protein is predicted to have a short luminal N-terminal domain, a single transmembrane (TM) spanning domain at the amino terminus that serves as an uncleaved signal peptide, and two cytoplasmic charged hydrophilic α-helices-interconnected by a flexible loop containing a highly conserved phosphorylated sequence (D52SGNES57)-at the carboxyl terminus [35]. Moreover, the TM was found to down-regulate the CD4 receptor [27], while the deletion of the highly negatively charged C-terminal end (with three aspartic residues in the last five amino acids) was observed to be detrimental to in vitro VpU-induced CD4 degradation [6].
VpU is associated with two primary functions during the HIV-1 life cycle. First, it contributes to viral-induced CD4 receptor down-regulation by mediating the proteasomal degradation of newly synthesized CD4 molecules in the endoplasmic reticulum. Secondly, it enhances the release of viral progeny from infected cells by antagonizing tetherin that is an interferon (IFN)-regulated host restriction factor which directly cross-links virions on the host cell surface [14]. Recently, a novel function of VpU related to stabilization of tumour suppressor p53 was reported: the modulation of the p53-stability correlates positively with apoptosis during late stages of HIV-1 infection [43]. These observations allow us to consider VpU to be an antagonist of the innate immune response to viral infection.
As with all other retroviruses, HIV-1 employs a variety of different overlapping reading frames and splicing events to express a large array of mRNAs (at least 40) and proteins (at least 15) from a single primary transcript. During HIV-1 viral replication, three classes of RNAs are produced: (ⅰ) the early doubly spliced approximately 2-kb transcripts encoding Tat, Rev, and Nef; (ⅱ) the late singly spliced mRNAs encoding Vif, Vpr, VpU, and Env products; and (ⅲ) the late approximately 9-kb unspliced mRNAs that are packaged into progeny virions as genomic RNA and which can also serve for the expression of Gag/Pol genes [12, 24]. Specifically, the viral envelope (Env) glycoprotein is translated from about 16 alternatively spliced 4 kb bicistronic mRNA Rev-dependent isoforms that all contain the upstream ORF for VpU. (Rev is a virally encoded sequence-specific RNA-binding protein that shuttles viral mRNAs between the nucleus and the cytoplasm). The biosynthesis of the Env polyprotein -starting about 160 nucleotides downstream from the VpU start codon -implies that the expression of VpU and Env from the same 4-kb spliced mRNA isoforms through leaky scanning translation are coordinated during HIV-1 infection [8, 26, 36].
HIV-1 entry into the host cell is mediated by the Env glycoproteins, gp120 and gp41: the initial binding of gp120 to the cellular CD4 receptor triggers conformational changes in gp120 that promote its following interaction with one of the chemokine co-receptors, usually CCR5 or CXCR4. HIV-1 strains can be phenotypically classified according to their ability to use the CCR5 and/or CXCR4 co-receptor: this binding is based upon the presence of selected amino acids in gp120 (specifically within the V3-loop, but also in other proteic regions) providing greater affinity to CCR5 or CXCR4, and therefore the viral tropism [11, 45].
In this light, the aim of the present study was to genetically characterize HIV-1 B-subtype gp120V3 and VpU sequences in terms of co-receptor usage and to define the association of amino acid changes within the V3 and the VpU regions according to CCR5 and/or CXCR4 usage.
HTML
-
A large cohort of 239 HIV-1 subtype-B sequences containing the specific genomic region from VpU to gp120 V3 domain, all retrieved from Los Alamos Database (one sequence per individual) [http://www. hiv.lanl.gov] were analyzed. All analyzed sequences have a pure phenotype and/or co-receptor determinations available (119 R5-and 120 X4-using viruses, respectively). Sequences were all retrieved from infected individuals at all stages of infection and collected from several countries representing all geographic continents (sampling years: 1983-2009). The drug treatment status for the individuals is not available in the Los Alamos Database, but some sequences were retrieved from ART naï ve patients (16 R5-and 2 X4-tropic viruses, respectively). As previously reported [11, 13], the multiple sequence alignments of VpU and V3 viral segments were performed by using ClustalX and Bioedit software packages [http://www.es.embnet.org http://www.mbio.ncsu.edu]. To confirm the HIV-1 subtype-B, published consensus sequences of pure HIV-1 subtypes (A, B, C, D, F1, F2, G, H, J, and K) were used [http://www.hiv.lanl.gov], and multi-aligned sequences were subjected to phylogenetic inference [11, 13]. To analyze VpU and V3 signatures, the frequency of all substitutions in the 81 VpU amino acids and 35 V3 amino acids were calculated. Fisher exact tests were used to determine the differences in frequency between the 2 groups of patients (infected with R5-or X4-using viruses, respectively). The Benjamini-Hochberg method was been used to identify results that were statistically significant in the presence of multiple-hypothesis testing [2]. A false discovery rate of 0.055 was used to determine statistical significance.
To identify significant patterns of pairwise associations between V3 and VpU amino acid changes, the phi coefficient (φ) and its statistical significance for each pair of mutations were calculated [11, 13, 40].
-
Consistent with previous observations, the first analysis was to confirm the classical V3 positions 11 and 25. The serine wild-type at position 11 (indicated as S11S) and the E25D mutation were significantly associated with R5-tropic viruses, while the S11KR and E25KRQ mutations were significantly associated with CXCR4 co-receptor usage [9, 15] (Fig. 1a).
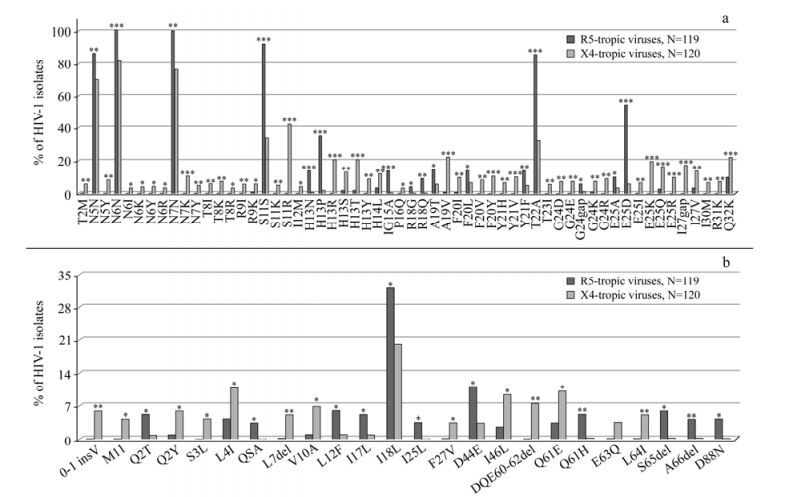
Figure 1. Frequencies of HIV-1 gp120V3 and VpU amino acid changes. Frequencies of V3 (panel "a") and VpU (panel "b") mutations in HIV-1 R5-tropic and HIV-1 X4-tropic isolates. The analysis was performed in sequences derived from 239 patients, 119 reported as R5-tropic and 120 reported as X4-tropic at phenotypic test. Statistically significant differences were assessed by chi-square tests of independence. P values were significant at a false-discovery rate of 0.05 following correction for multiple tests. *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
By evaluating the phenotypic dataset of V3-loop sequences, we tested our previous genotypic results [11]: 8 out of 9 V3 previously predicted mutations (89%) were present with a prevalence significantly higher in R5-than in X4-tropic viruses (the known mutations were E25D, and H13N, H13P, G 15A, R18Q, F 20L, Y 21F and T 22A) (P < 0.05) (Fig.1a). In addition, the following V3 signatures, R 18G, A19T, G24gap, and E 25A, were found to be significantly associated with R5-tropic viruses (P < 0.05) (Fig. 1a).
Moreover, 28 out of 33 previously predicted amino acid changes (85%) were significantly more prevalent in X4-than in R5-tropic viruses (the known mutations were the classical S11KR and E25KRQ, and T2M, N6Y, N7K, T8I, R9K, I12M, H13RSTY, P16Q, A19V, F20IVY, Y21HV, G24EKR, I27V, R31K, Q32K) (P < 0.05) (Fig. 1a). The following V3 signatures, N5Y, N6IKR, N7Y, T8KR, R9I, I14L, T23I, G24D, E25I, I27gap, and I30M, were found to be significantly associated with X4-tropic viruses (P < 0.05) (Fig. 1a).
The conserved V3 region encompassing the residues 4 to 7 (P4-N5-N6-N7) has been demonstrated to interact with the CCR5 co-receptor: the binding of this co-receptor is blocked when N7 is replaced by a charged amino acid [20]. As previously observed [11], the phenotypic dataset in the present study indicates that the mutation N7K has been found only in X4-tropic viruses (prevalence:10.8%; P = 0.0002) (Fig. 1a).
By analyzing the VpU sequences, 23 out of 81 VpU positions were found to be significantly associated with different co-receptor usage for the first time (P < 0.05) (Fig. 1b). In particular, 11 VpU signatures whose prevalence was significantly higher in R5-than in X4-tropic viruses, were identified: 7 of them had a prevalence > 5% in R5-predicted viruses (Q2T, L12F, I17L, I18L, D44E, Q61H, and S65del) (Fig. 1b). Conversely, 13 substitutions whose prevalence was significantly higher in X4-than in R5-viruses were identified, suggesting their association with CXCR4-usage. Among the 13 substitutions, 9 signatures had a prevalence > 5% in X4-tropic viruses (0-1insV, Q2Y, L4I, L7del, V10A, I46L, DQE60-62del, Q61E, and L64I) (Fig. 1b).
Mutation of the VpU start codon occurs at low frequency during PBMC culture of HIV-1 isolates in vitro (1.2%; 12 of 967) [10]: recently, it has been shown that the HIV-1 YU-2 clone (highly macrophage-tropic) carries a mutated VpU start-codon. A low frequency (nearly 5%) of VpU sequences in the HIV sequence database contain a mutated AUG translation start codon [10]. This mutation failed to confer macrophage replication [33]. Our results are consistent with previous observations: 0-1insV and M1I are signatures retrieved only in X4-tropic viruses, with 5.8% and 4.2% of prevalence, respectively (Fig. 1b).
Several residues associated with different co-receptor usage reside within the VpU TM domain (amino acids 6-28) (L7del, V10A, L12F, I17L, I18L, I25L, and F27V): the fact that all these signatures are localized in the unexposed domain of the protein suggests that they may act as a scaffold in order to maintain the stability of the phospholipids-protein interaction. Notably, the changes are biochemically hydrophobic (non-polar) amino acid substitutions that are not expected to perturb the protein significantly (and so might be conveniently classified as good conservative substitutions): this supports the conservation of the TM domain functionality. Biochemical and biophysical evidence have suggested that this is critical for the VpU oligomerization (most favourably as a pentameric form) [21], as well as for down-regulating the CD4 receptor [27]. Moreover, a single mutation at VpU position 18 by a histidine has been shown to make the virus infection sensitive to the M2 channel blocker Adamantanes [19, 31]. In our dataset, the I18H mutation was absent, while I18L was the more prevalent mutation retrieved in all sequences (31.9% in R5-and 20.0% X4-tropic virus, respectively), followed by the polymorphism I18V (1.7% in R5-and 2.5% X4-tropic virus, respectively). Thus, only conservative amino acid changes in this position were found: the local modification of the primary structure plays an important role in secondary and tertiary structures of VpU TM in lipid bilayers and may affect its ability to interact with channel blockers.
Conversely, for the five minor variants observed in the predicted short luminal N-terminal domain (Q2T, Q2Y, S3L, L4I, and Q5A), with the exception of L4I, range of amino acid changes is relatively un conserved (different biochemical functionality) [42]: semi-conservative substitutions (Q2T and Q5A) and non-conservative changes (Q2Y and S3L) were observed. Although the accessory proteins are in general not necessary for viral propagation in tissue culture, several experimental observations suggest that their role in vivo is very important [14, 24]. This would suggest that, the short luminal peptide probably has no essential and mandatory role in biochemical conservation and infectivity.
Previous analysis of the VpU cytosolic domain have revealed the presence of two putative trafficking signals (amino acid regions: 29-34 and 63-68) that harbour a degree of amino acid variation among different subtypes [32]. For the first signal region (amino acids 29-34), in the present study the analysis of the prevalence mutations has evidenced a similar variability between R5-and X4-tropic viruses (Fig. 1b). Likewise, for the flexible loop D52SGNES57 at the VpU carboxyl terminus, that interconnects the two cytoplasmic charged hydrophilic α-helices, we did not observe the specific tropic-signatures.
On the other hand, in the first charged hydrophilic α-helix the D44E (in R5-tropic viruses) and the I46L (in X4-tropic viruses) were found (Fig. 1b), while several tropic-related positions were found in the second α-helix containing the other trafficking domain (Q61H, S65del, and A66del, for the R5-tropic viruses, and DQE60-62del, Q61E, E63Q, and L64I, for the X4-tropic viruses) (Fig. 1b).
Finally, only the D80N R5-tropic signature was observed in the C-terminal end of VpU (amino acids 76-81) (Fig. 1b). Interestingly, the deletions in several regions of VpU were observed both in R5-and in X4-tropic viruses (residue position 7, 60-62, 65 and 66): on the contrary, deletion of the C-terminal residues would not be allowed because it would be deleterious to the virus, probably resulting in the loss of interaction with CD4 and the inability to induce CD4 degradation [29].
Although the second part of the encoding VpU sequence overlaps the 5' end signal sequence of HIV-1 Env (starting from the codon of amino acid residue N55VpU) [16], only a third of "tropic-signatures" within this region were observed (Fig. 1b). Interestingly, this overlapping region was found to be related to different viral effects. Michalski et al. have shown that the signal peptide of HIV-1 Env itself has a direct role in cellular cytotoxicity and the triggering of cell death pathways [30].
At the same time, we underline that various classes of small non-coding RNAs (sncRNAs) are important regulators of gene expression across the divergent types of the organisms, with virally encoded sncRNAs (particularly those of RNA viruses) expressed at very low levels. Althaus and his co-workers have captured almost 900 HIV-1 sncRNAs, some of them distinguishable retrieved in the VpU, Env and VpU/Env overlapping viral regions [1]. In this last cited overlapping region a moderate variability related to the viral tropism was allowed. The HIV-1 encoded sncRNAs vary in length and in their locations on the viral genome and they may have the potential to play roles in HIV-1 replication [1]. Thus, we wanted to emphasize these aspects in relation to the results of VpU association with the first step of viral cycle (the binding between viral envelope proteins and the host cellular surface).
Statistically-significant correlations among V3 and VpU signatures in HIV-1 clade B were identified for the first time (Table 1). Some of these correlations involved the classical V3 positions 11 and 25 (Table 1). Specifically, the E25KRQV3 mutations showed positive correlations with several VpU signatures (0-1insV, I46L, and 60-62del). Similarly, S11KRV3 mutations showed positive correlations with I46LVpU and 60-62delVpU signatures (Table 1). All these amino acid variants correlate with CXCR4-usage. Conversely, S11SV3 established a positive correlation with only one VpU amino acid change, Q61H (P=0.041; φ=0.14), localized in the second trafficking domain contained within the second cytosolic charged hydrophilic α-helix (Table 1). Of note, S11SV3 was found in 100% of patients with Q61HVpU: the association of these two amino acid residues has always shown codon cau for the Q61HVpU and codon agu for S11SV3, further supporting that these signatures are strongly correlated with each other both at the nucleotide and amino acid level.

Table 1. Novel VpU signatures significantly associated with specific gp120V3 amino acid changes
However, the classical E25DV3 R5-tropic mutation was not established to have a statistically significant correlation with VpU amino acid residues: on the other hand, the classically strong association with S11SV3 was confirmed (P = 3.54e-11; φ = 0.41).
Among the positive correlations between V3 and VpU signatures associated with CXCR4-usage, strong correlations were observed for I30MV3 with I46LVpU (P = 3.61e-9; φ = 0.65) and with 60-62delVpU (P = 3.86e-11; φ = 0.82) (Table 1). Of note, I30MV3 was found in 50% of patients with I46LVpU, and in 77.8% of patients with 60-62delVpU: in the co-variation frequency the I46LVpU mutation has shown to always have the codon cua (for the I30MV3 substitution, it is known that the metionine was encoded by only one codon), further supporting the case that these substitutions are strongly correlated with each other both at the nucleotide and amino acid level (Table 1). Moreover, it has been recently shown that I30V3 is critically important for efficient HIV-1 entry into macrophages [5, 17]: the M30I in non-M-tropic CXCR4-tropic Envs enhances macrophage entry via CXCR4. These reports support a critical role for I30 in promoting efficient X4-mediated HIV-1 entry into macrophages [5, 17].
Another positive correlation between V3 and VpU signatures associated with CXCR4-tropic was the E25IV3 with V10AVpU (P = 0.032; φ = 0.21) (Table 1). In an in vivo study new V3-genetic mutations modulating co-receptor usage were identified [41], and the E25IV3 was listed as a significantly associated minor variant with CXCR4-tropic (with low prevalence). In the present analysis this preliminary observation was confirmed, with a prevalence of 6.7% in X4-vs 0% in R5-tropic viruses, respectively (P=0.004).
In the V3-loop it has been shown that the secondary structure of the GPGX crown (at positions 15-18) is crucial in modulation of HIV-1 subtype co-receptor usage. This motif forms a proteic β-turn that binds to the extracellular loops of the co-receptor [23]. Among the positive correlations between V3 and VpU mutations associated with CCR5-usage, a correlation for D80NVpU with a R18QV3 substitution was observed (P = 0.018; φ = 0.25). The V3 position 18 -along with position 20 -resides in a motif shown to be involved in the binding with two specific glycosphingolipids (GSLs): galactosylceramide and sphingomyelin. This binding has been shown to mediate the attachment of HIV-1 to plasma membrane microdomains (rafts). Several works suggest that GSLs are involved in the entry of a broad range of HIV-1 isolates into cell lines expressing CD4, CCR5 and/or CXCR4, and the GSL depletion blocked subsequent viral fusion and infection [44].
The VpU amino acid change L12F, localized in the first part of the TM domain, also established a positive correlation with two V3 mutations, E25A (P = 0.012; φ= 0.23) and T22A (P = 0.015; φ = 0.15) (Table 1). Of note, T22AV3 was found in 100% of patients with L12FVpU: in the covariation frequency these two mutations has shown to always have the codon gca (for the T22AV3) and the codon uuu (for L12FVpU), further underlining that these mutations are strongly correlated with each other both at the nucleotide and amino acid level.
The correlation of signatures in V3 and VpU was also confirmed by hierarchical clustering analysis [11, 13, 40]. In particular, the topology of the dendrogram suggests the existence of a three sub-clusters associated with R5-tropism composed of five V3 mutations and four VpU signatures: E25DV3 and V10AVpU, (bootstrap=0.71); 0-1insVVpU, H13RV3, I46LVpU, I30MV3, and 60-62delVpU (bootstrap = 0.88); S11K/RV3 and E25DV3 (bootstrap =0.64). These sub-clusters were part of a large R5-cluster (bootstrap = 0.97) (Fig. 2). Similarly, another large cluster was found associated with X4-tropism (bootstrap =0.94). This involves three sub-clusters: D44EVpU, R18QV3 and D80NVpU, (bootstrap = 0.63); E25AV3 and L12FVpU, (bootstrap = 0.70); Q61HVpU, T22AV3, S11SV3, E25DV3, and H13N/PV3 (bootstrap = 1) (Fig. 2).
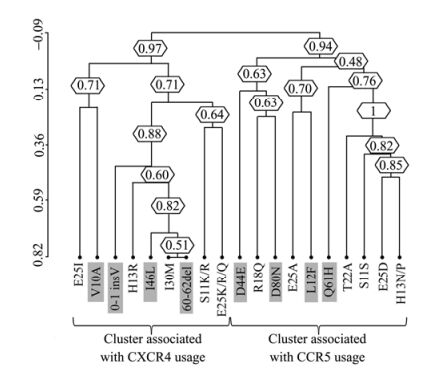
Figure 2. Clusters of correlated V3 and VpU signatures. Dendrogram obtained from average linkage hierarchical agglomerative clustering, showing significant clusters involving V3 and VpU (gray box) signatures. The length of branches reflects distances between mutations in the original distance matrix. Boostrap values, indicating the significance of clusters, are reported in the boxes. The analysis was performed in sequences derived from 239 patients, 119 reported as R5-tropic and 120 reported as X4-tropic at phenotypic test.
-
In addition to co-receptor usage, there may be many different reasons for the correlation among V3 and VpU amino acid changes. Possibly, these associations may have an impact on the HIV-1 pathogenesis. It is known that the CXCR4 phenotype has been associated with progression and increased severity of HIV-1 disease. Recently, Marchal and his co-workers highlighted a novel functional link between the VpU and caspase-dependent apoptosis via the activation of the c-Jun N-terminal Kinas pathway [28]. Moreover, VpU-deficient virions have been tightly associated with the cell surface, indicating a possibility that VpU can convert the mode of virus transmission [22]. Consistently, it has been also found that VpU indirectly enhances retroviral release through modification of the cellular environment (numerous reports have shown the high complexity of the relationships between VpU and cellular proteins of the host) [18]. Hence, some of the VpU signatures observed here could also be linked with the phenomena listed above.
In addition, Stephens and his co-workers have suggested that removal of the VpU sequence upstream of Env resulted in enhanced Env precursor synthesis, with a viral capacity of low pathogenic phenotype for pig-tailed macaques [38], while Casella et al. have observed the capacity of VpU to increase the viral susceptibility in infected cells to Fas killing [4]. Moreover, other studies have already indicated the importance of VpU for virus replication in macrophages [10, 25, 34]. These other details underline the complexity of the HIV-1 life cycle via viral factors and expression of additional proteins (regulatory and accessory) that can regulate its virulence and pathogenesis. In our analysis, specific variable residues were shown to be correlated with primary tropic amino acid signatures in the gp120-V3-loop that strengthens the case for a role of VpU that is more than a "simple" accessory protein.
For the sake of completeness, previous studies have shown that the VpU and Env proteins are translated from different reading frames from the same messenger RNA (mRNA) species [36]. Thus, the unique structure of these mRNAs suggests that the upstream VpU sequences prior to the Env could condition the expression of the Env polyprotein. Occasionally, VpU tropic-properties have described in the past [10, 37], generally associated with intracellular location of this protein and consequently not with the virus entry steps. Nevertheless, the co-presence of Env and VpU ORFs in the same mRNAs (in late phases of viral infection) [24] should suggest a closer relationship between VpU and viral tropism. The selection in the gp120-V3 domain of mainly R5-and X4-tropic mutations could determine a secondary forced selection of mutations in the VpU sequence. Specifically, since HIV replication depends upon the stability of its RNA genome and its single spliced transcripts within the infected cell, the conservation of some positions could be a direct consequence of the need for the virus to maintain the proper folding of genomic RNA into highly stable RNA paired stems. Hence, after the discovery of VpU as a functional gene [7, 39], we now hypothesize that its gene product may have an important function not only in regulating virus release, but also has significant association with different co-receptor usage and specific V3 mutations.
In summary, this study shows that specific VpU substitutions are significantly associated with different co-receptor usage and with specific V3 mutations, both at the nucleotide and amino acid level. Specific in vitro studies are needed to exclude an hypothetical statistical false positive result, to confirm that these VpU mutations contribute directly to co-receptor usage and to establish the specific and precise utility of this information.













 DownLoad:
DownLoad: