-
Synechococcus, some of which differ genetically and physiologically and display different environmental distribution patterns, are found in high concentrations in coastal waters (Scanlan D J, 2002; Waterbury J B, 1986). It has been estimated that in coastal waters, 80% of Synechococcus cells may encounter infectious cyanophage particles each day (Suttle C A, 1994). Although many Synechococcus isolated from coastal waters are resistant to cyanophages, a large number of cyanophages have been successfully isolated from different coastal environments (Suttle C A, 1994; Waterbury J B, 1993).
Cyanophages are viruses that can infect Synechococcus sp. and other prokaryotic algae. The isolation and characterization of cyanophages infecting marine Synechococcus strains began in the 1990s (Suttle C A, 1993; Waterbury J B, 1993; Wilson W H, 1993). Five marine cyanophages propagated on Synechococcus sp. strain WH7803 were isolated by Waterbury and Valois from different oceanographic areas, including open ocean and coastal waters (Waterbuty J B, 1986; Waterbury J B, 1993). Representatives of all three families of tailed phages were found among the seven Synechococcus phages isolated by Suttle, Chan and Wilson (Suttle C A, 1993; Wilson W H, 1993). The diversity of cyanophages in the environment is substantial, and many new cyanophages have been discovered over the past decade (Prangishvili D, 2006; Suttle C A, 2007; Van Etten J L, 2010). However, metagenomic data show that more than 70% of the genes in the oceanic viral fraction cannot be associated with known viruses, indicating that a substantial amount of viral diversity remains to be identified (Angly F E, 2006; Bench S R, 2007; Suttle C A, 2007).
Cyanophages infecting Synechococcus are ubiquitous in marine environments and are highly abundant in coastal waters, with concentrations of up to 108 cyanophages per liter (Suttle C A, 1993; Suttle C A, 1994; Waterbury J B, 1993). However, no previous reports have documented marine Synechococcus cyanophages in China. The purpose of this study was to isolate a cyanophage-Synechococcus system from the East China Sea; this goal is fundamental for studying phage-host interaction in the Chinese coastal ecosystem.
HTML
-
Surface seawater samples were collected from the coastal waters of the East China Sea (33°44'38″N, 122°25'44″E) in August 2011 at a depth of 1 m. The water samples for the isolation of cyanophages were concentrated by the ultrafiltration method described by Chen et al. and stored in the dark at 4 ℃ until use(Chen F et al, 1996). The water samples for the isolation of Synechococcus were filtered through a 1.0-mm filter and stored at 15 ℃ until use.
-
The filtered samples were enriched with sterilized artificial seawater medium (ASW) and incubated under a constant illumination of approximately 25 μE m-2 s-1 at 20 ℃. After visible growth (based on color and turbidity) was evident in the enrichments, clonal isolates were obtained by repeated cycles (repeated five times) of colony isolation using a serial dilution method described by Throndsen and Rippka (Rippka R, 1988; Throdson J, 1969). The resulting isolate SJ01 was clonal and unialgal.
-
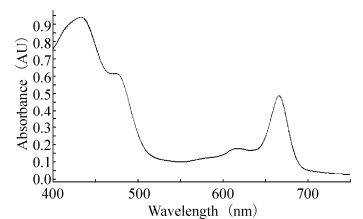
Cultures in the exponential to early stationary phase were used for recording in vivo absorption spectra. An aliquot of the culture was transferred to a cuvette, and the in vivo absorption spectrum was measured from 400 to 750 nm using a Scinco S-3100 UV-VIS Spectrophotometer equipped with LabPro Plus Software. The A495/A545 ratio is a measure of the amount of PUB (phycourobilin) relative to that of PEB (phycoerythrin) (Rocap G, 2002). The experiment was performed in triplicate.
Chlorophyll was estimated using a previously described method (Becker W E, 1994). The cells were resuspended in 90% methanol and allowed to stand in the dark for 24 hours (Becker W E, 1994). The suspension was cleared by centrifugation, and chlorophyll was estimated spectrophotometrically using the following equations: Chlorophyll a (μg L-1) = (13.95×A665) - (6.88×A650), and Chlorophyll b (μg L-1) = (24.96×A650) - (7.32×A665)(Rocap G, 2002), where A650 and A665 are the absorbance values of the sample at 650 and 665 nm, respectively. The experiment was performed in triplicate.
-
DNA was prepared from exponential-phase culture using a method modified from that described by Tillett and Neilan (Tillett D and Neilan B A, 2000). The extracted DNA (2 μL) was added to a 48 μL PCR mixture containing Taq DNA polymerase assay buffer (50 mmol/L KCl, 20 mmol/L Tris-HCl, pH 8.4), 5.0 mmol/L MgCl2, 200 μmol/L deoxyribonucleoside triphosphate, 0.25 μmol/L each of the primers, and 2.0 U of Taq DNA polymerase (TaKaRa). The PCR primers that were used to amplify the 16S rRNA, ITS, and psbA genes of Synechococcus sp. SJ01 are shown in Table 1. The negative controls contained all of the above reagents, but sterile water was used as the template. The amplification products were verified by electrophoresis and then sequenced.
Species Primer Sequence Source Synechococcus SJ01 16S rRNA 27F1 5′-AGAGTTTGATCMTGGCTCAG-3′ Neilan B A, 1997 409R 5′-TTACAA(C/T)CCAA(G/A)(G/A)(G/A)CCTTCC TCCC-3′ ITS 16S-1247f 5′-CGTACTACAATGCTACGG-3′ Rocap G, 2002 23S-1608r 5′-CYACCTGTGTCGGTTT-3′ psbA psbA-1 5′-AACATCATYTCWGGTGCWGT-3′ Lindell D, 2004 psbA-2 5′-TCGTGCATTACTTCCATACC-3′ Cyanophage S-SJ2 g20 CPS1 5′-GTAG[T/A]ATTTTCTACATTGA[C/T]GTTGG-3′ Sullivan M B, 2008 CPS2 5′-GGTA[G/A]CCAGAAATC[C/T]TC[C/A]AGCAT-3′ psbA psbA-1F 5′-AACATCATYTCWGGTGCWGT-3′ Huang S, 2012 psbA-1R 5′-TCGTGCATTACTTCCATACC-3′ DNApol 90Fa 5′-GAYACIYTIRYIYTITCIMG-3′ Huang S, 2012 355Ra 5′-GGIAYYTGIGCIARRTTIGG-3′ Table 1. Primers used in this study
BLAST searches of the GenBank database (www.ncbi. nlm.nih.gov) were performed to identify closely related sequences. Neighbor-joining (NJ) trees were generated using the MEGA 4.0 program (http://www.megasoftware.net/) for all sequences, which included selected representative sequences from GenBank following alignment using ClustalX2. Bootstrap values were obtained with 1000 resamplings, and clades with bootstrap values greater than 50% were shown on the nodes of the branches.
-
Synechococcus phages were isolated with liquid dilution cultures rather than from plaques on solid media because lawn formation is erratic for many strains belonging to Synechococcus marine cluster A (Waterbury J B, 1993). Serial dilutions of the concentrated seawater samples were added to exponentially growing cultures of the host suspended in ASW growth medium in test tubes. The phage-host suspensions were incubated at 20 ℃ for 1 h with occasional agitation to allow the adsorption of the cyanophage. These tubes were incubated in constant illumination (approximately 25 μE m-2 s-1) at 20 ℃ and were monitored daily for cell growth or lysis. The controls contained host cells but no seawater dilutions. The lysis of the host cells was usually observed for up to five weeks. Putative cyanophage lysates obtained by this method were passed through a 0.22 μm pore size filter, and the filtrate was stored at 4 ℃ in the dark. The purified cyanophage filtrates were then obtained with three successive serial dilutions in the liquid medium.
-
Phage purification was performed by sucrose density gradient centrifugation as described previously with the following modifications (Vidaver, 1973): both DNA and RNA were digested with 1 μg/mL Dnase Ⅰ and Rnase A (TaKaRa); and five sucrose steps of 20%, 30%, 40%, 50%, and 60% were used. A visible band locating between the 40% and 50% steps was collected and washed to obtain highly purified phage particles.
-
An ultra-thin section was used to observe the ultrastructure of Synechococcus sp. SJ01. One milliliter Synechococcus sp. SJ01 was centrifuged at 6000 g for 10 min, and 0.2 mL 2.5% glutaraldehyde was then added to the precipitate for postfixation. Fixation was performed by immersing the sample in 1% osmic acid for 4 h. The sample was then centrifuged at 6000 g for 5 min. PBS was added to the precipitate for washing, and the centrifugation and washing were repeated three times. Dehydration was performed by immersing the sample in 50% to 100% alcohol. The sample was then embedded in Spurr resin (ERL-4206). Ultra-thin sections were stained with uranyl acetate and lead citrate and then observed using a Tecnai G2 TEM.
A total of 20 μL of sucrose density gradient purified phage concentrate was transferred to 200 mesh formvar carbon-coated copper grids and then negatively stained with 2% sodium phosphotungstate (pH 7.0). The grids were viewed using a HITACHI 3H-7000FA TEM.
-
To estimate the size of the S-SJ2 genome, DNA was prepared from sucrose density gradient purified phages following the method of Wilson et al (Wilson, 1993). The DNA that was extracted from purified cyanophage was digested with the following restriction endonucleases: Hind Ⅲ, BamH Ⅰ, EcoR Ⅰ, EcoR Ⅴ, Pst Ⅰ, and Dpn Ⅰ. Extracted DNA (2 μL) was added to a 48 μL PCR mixture containing Taq DNA polymerase assay buffer (50 mmol/L KCl, 20 mmol/L Tris-HCl, pH 8.4), 5.0 mmol/L MgCl2, 200 μmol/L deoxyribonucleoside triphosphate, 0.25 μmol/L each of the primers, and 2.0 U of Taq DNA polymerase (TaKaRa). The PCR primers that were used to amplify the g20, psbA, and DNApol genes of cyanophage S-SJ2 are shown in Table 1. Negative controls contained all of the reagents, but sterile water was used as the template. The amplification products were verified by electrophoresis.
Location and sampling
Synechococcus isolation and growth conditions
In vivo absorption spectra and pigment measurements of Synechococcus
DNA extraction, PCR amplification, sequencing and phylogenetic analyses of SJ01
Phage isolation
Phage purification
TEM observation of Synechococcus and cyanophage
DNA extraction, PCR amplification and restriction enzyme digestion of the phage S-SJ2 genome
-
The cells of Synechococcus sp. strain SJ01 averaged ca. 0.8 μm in width and 1.5 μm in length and were spherical to oval in shape (Fig. 1). No well-defined sheath layer or gas vacuoles were observed. Photosynthetic thylakoids of SJ01 were peripheral and oriented parallel to the cytoplasmic membrane. Several carboxysomes were observed in the central portion of the cells. The dividing cells showed transverse binary fission.
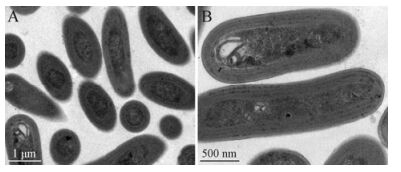
Figure 1. Transmission electron micrographs of ultrathin sections of cells from strain SJ01. Scale bar, (A) 1 μm, (B) 0.5 μm. The cells of Synechococcus sp. strain SJ01 averaged ca. 0.8 μm in width and 1.5 μm in length and were spherical to oval in shape. Photosynthetic thylakoids were peripheral and oriented parallel to the cytoplasmic membrane.
In vivo absorption spectra indicated that this Synechococcus SJ01 absorbs light efficiently in the blue region, with two prominent peaks at 440 nm (chl a) and 492 nm (primarily due to PUB and carotenoids) (Fig. 2). On the basis of the PUB/PEB ratios, the Synechococcus sp. strain SJ01 was remarkably similar to the motile strain WH8102, which possessed phycoerythrin (PE) as the major light-harvesting pigment (Waterbury J B, 1986). Moreover, the chl b/a ratios of the Synechococcus sp. strain SJ01 was only 0.17, which differed from the chl b/a ratios of Prochlorococcus strains, which ranged from 0.20 to 1.41 (Rocap G, 2002).
-
All of the PCR amplifications yielded single-band products, and the lengths of the 16S rRNA, ITS and psbA gene sequences were ≈400 bp, ≈1200 bp, and ≈750 bp, respectively. The Synechococcus isolate SJ01 is phylogenetically closely related to Synechococcus sp. WH8102 (Accession: BX569694.1), and the sequence identities between them were 99%, 94%, and 91% based on their 16S rRNA, ITS, and psbA gene sequences, respectively. In phylogenetic analyses of this strain, based on the 16S rRNA, ITS, and psbA sequences, SJ01 was always closely related to the members of Marine A Synechococcus clade Ⅲ (Fig. 3). The sequencing results of 16S rRNA, ITS, and psbA sequences of Synechococcus SJ01 are shown in Supplementary material S1.
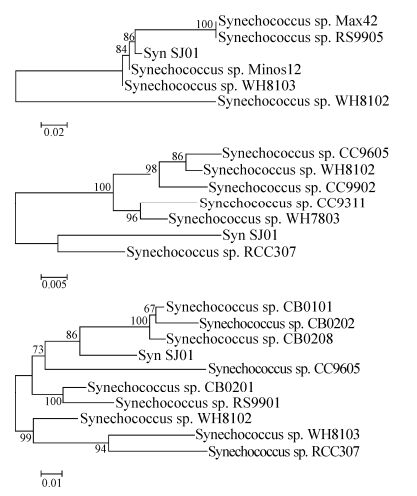
Figure 3. Neighbor-joining phylogenetic tree of 16S rRNA (A), ITS (B), and psbA (C) genes from marine Synechococcus strains (with Jukes-Cantor correction). The tree was constructed from an alignment using Clustal X2. The bootstrap values were obtained through 1000 repetitions. The sequences were extracted from BLAST searches of GenBank.
-
Using liquid dilution cultures and sucrose density gradient centrifugation, we isolated and purified a marine Synechococcus siphovirus, S-SJ2, infecting strain SJ01. The morphology of negatively stained S-SJ2, as observed with TEM, revealed an icosahedral capsid that was ≈68 nm in diameter and exhibited a long tail (≈280 nm long and ≈20 nm in diameter) (Fig. 4). S-SJ2 was morphologically similar to S-CBS4, a siphovirus infecting marine Synechococcus CB0101, which also has an isometric head (≈72 nm) and a long flexible tail (≈200 nm) (Huang S, 2012).
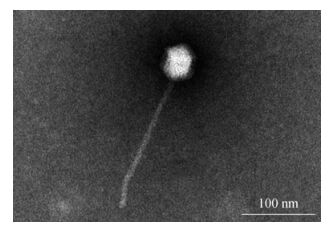
Figure 4. Purified S-SJ2 particles visualized using a negative staining method. Scale bar, 100 nm. The phage particles were purified by liquid dilution cultures and sucrose density gradient centrifugation. The sample was negatively stained with 2% sodium phosphotungstate (pH 7.0) and observed at a magnification of ×100000.
-
Agarose gel electrophoresis of the nucleic acid extracted from purified cyanophage S-SJ2 showed that the genome is less than 23 kb in length (Supplemental material, Figure S1). DNA extracted from purified S-SJ2 could not be restricted with the restriction endonucleases HindⅢ, BamHⅠ, EcoRⅠ, EcoRⅤ, PstⅠ, and DpnⅠ. The g20, psbA and DNApol sequences from S-SJ2 could not be amplified by any of the primer sets in this study. This result was consistent with observations made by Huang et al (Huang S, 2012).
Morphology and pigmentation of the strain
Molecular identification of the strain
The morphology of the cyanophage
Genome features of cyanophage S-SJ2
-
The cyanobacterial picoplankton SJ01 isolated from a coastal area of the East China Sea was identified as Synechococcus sp. based on morphological features and 16S rRNA, ITS, and psbA sequence data. The strain was very similar to Synechococcus sp. WH8102, a member of clade Ⅲ of the marine cluster A (MC-A) group. This group has been isolated from the euphotic zone from both open-oceans and coastal waters and has an elevated salt requirement for growth (Fuller N J, 2003). In contrast with the open-ocean strains, certain coastal strains of clade Ⅲ of marine cluster A (e.g., the SJ01 and WH8103 strains) present a very high PUB. Their presence in the coastal environment may be more representative of physical oceanographic patterns (Ong L J, 1984; Vera T, 2009). Their occurrence in late summer coincides with periods of strong stratification (Palacios D M, 2004). Partensky et al. have stated that the vertical distributions of Prochlorococcus and Synechococcus populations tend to exhibit certain general features (Partensky F J, 1999). Typically, Synechococcus populations are approximately 10-fold less abundant than Prochlorococcus populations except in coastal waters (Mann N H, 2003). Due to its photosynthetic lifestyle and sheer abundance, Synechococcus contributes 20% of the primary productivity in certain coastal environments (Li W K W, 1994).
Three families of cyanophages, Myoviridae, Siphoviridae, and Podoviridae, have been reported. According to its morphological characteristics, S-SJ2 was very similar to S-CBS4, a siphovirus infecting marine Synechococcus CB0101, which was isolated from Chesapeake Bay (Huang S, 2012). All six of the restriction endonucleases tested failed to digest cyanophage DNA into clearly definable band patterns. This result was consistent with observations made by Wilson et al. (Wilson W H, 1993) and Lu et al (Lu J, 2001). Many cyanophages or bacteriophages that are resistant to digestion by certain endonucleases have been described previously (Lu J, 2001). This resistance to endonuclease digestion could result from either an absence of restriction sites or, more likely, the presence of modified bases, such as those resulting from the methylation of cyanophage DNA. To date, the genome sequences of 28 cyanophages (17 myoviruses, 6 podoviruses, and 5 siphoviruses) isolated from marine ecosystems have been reported (Huang S, 2012). Based on different gene markers, such as the g20 gene encoding viral capsid assembly protein for cyanomyovirus, the viral DNA polymerase gene for cyanobacterial podoviruses, as well as the core photosystem Ⅱ psbA gene for both of these viruses, an extensive genetic diversity of cyanobacterial myoviruses and podoviruses has been found in various marine ecosystems (Huang S, 2012). However, the highly variable genomes of cyanobacterial siphoviruses suggest that it is impossible to identify a specific gene marker for this group of cyanophages (Huang S, 2012). The phage family Siphoviridae often contains temperate members with the capability of integrating into the host genome or entering a lysogenic lifestyle. For this reason, it is difficult to determine the lytic cycle and burst size of siphovirus S-SJ2 (Huang S, 2012). Like most previously reported unidentified cyanophage isolates, S-SJ2 was isolated without complete genome sequencing and identification, despite the fact that marine cyanobacteria have been used to isolate phages for two decades. (Wang K, 2011). The discovery of a novel cyanophage isolated on marine cyanobacteria substantially increases the likelihood that marine environments harbor many more novel phage types, especially those found on local cyanobacteria hosts that we are capable of culturing (Sabehi G, 2011). Given the dominance of Synechococcus-phage systems, it is unsurprising that many recent studies have found that phage-bacteria interactions strongly influence global biogeochemical cycles (Weitz J S, 2013).
Cyanophage-Synechococcus systems have been described from diverse habitats worldwide, including coastal and open-ocean environments. Since the early 1990s, more than 350 cyanophages infecting Synechococcus have been isolated, and 210 of these isolates have been further characterized (Marston M F, 2003; Wilson W H, 1993). Due to the importance of Synechococcus-phage interaction in the coastal ecosystem, the isolation and characterization of the Synechococcus-phage system has become an important topic in aquatic virology and marine environmental research (Singh P, 2012). Our study describes the first isolation of a cyanophage-Synechococcus system from the coastal ocean in China. The system shares many properties with other marine cyanophage-Synechococcus isolates, and additional details remain to be investigated. However, the successful establishment of a culturable cyanophage-Synechococcus system from the coastal ocean of China would be helpful for further research into phage-host interaction in the Chinese coastal ecosystem.
-
We thank Shichang Liu and Tao Wang for assistance in gathering samples and other technical assistance. This study was financially supported by NSFC (National Science Foundation of China, no: 31200385) and Science and technology project of Hubei Province (no: 2006AA305A04).
-
Yan Zhang carried out all the experiments. Min Xu performed the genetic analysis. Yijun Zhao provided finical and equipment support. Kai Cheng conceived and designed.
-
The supplementary materials are available on the website of Virologica Sinica: http://www.virosin.org.













 DownLoad:
DownLoad: