HTML
-
The sweet potato (Ipomoea batatas) is an important root crop that is ranked as the seventh in production worldwide. It is vegetatively propagated and therefore prone to virus accumulation. More than 20 viruses have been identified and described in sweet potatoes, including sweepoviruses (Valverde R A, et al., 2007).
Sweepoviruses, named for sweet potato begomoviruses, are mo nopartite (no DNA-B, alphasatellite or betasatellite component has been identified), and phylogenetically distinct from the Old and New World begomovirus groups (Fauquet C M, et al., 2003). Sweepoviruses infect plants of the genus Ipomoea worldwide, and have been found in the mainland of China (Luan Y S, et al., 2007; Bi H, et al., 2012)and Taiwan (Li R H, et al., 2004), Japan (Onuki M, et al, 1998), Israel (Cohen J, et al., 1997), Peru (Fuentes S, et al., 2003), Kenya (Miano D W, et al., 2006), Italy (Briddon R W, et al., 2006), Spain (Lozano G, et al., 2009), the USA (Zhang S C, et al., 2011), Uganda (Wasswa P, et al., 2011) and Brazil (Albuquerque L C, et al., 2012). Several reports have indicated substantial yield losses in some sweet potato cultivars due to infection by sweepoviruses (Clark C A, et al., 2006; Ling K S, et al., 2010). To date, most studies of sweepoviruses have been restricted to genomic cloning and sequence analysis. However, the infectivity of cloned sweet potato leaf curl Lanzarote virus (SPLCLaV) in sweet potato plants has been demonstrated using syringe infiltration (Trenado H P, et al., 2011). There are no reports of the successful infection of sweet potato plants by a sweepovirus clone using vacuum infiltration.
In the current study, vacuum infiltration was used to deliver an infectious clone of the sweet potato leaf curl virus-Jiangsu (SPLCV-JS), a sweepovirus recently identified in China (Bi H, et al., 2012), into Xushu 22, one of the most popular sweet potato cultivars in China.
-
Virus-free in vitro plantlets of the sweet potato cultivar Xushu 22 were subcultured on Murashige and Skoog (MS) basal medium (Murashige T, et al., 1962), which was supplemented with 2% sucrose and solidified with 0.3% Gelrite (pH 5.8) and cultivated in a growth chamber at 26℃ under a 16 h/8 h photoperiod with light intensity of approximately 60 μmol m-2s-1. Plantlets having 3-4 leaves were transferred into liquid MS basal medium in a jar and put in a humidor under the same culture conditions as described above for a week before vacuum infiltration. After infiltration, the plants were returned to the humidor and grown at 21±2℃ in a chamber with a 16 h/8 h photoperiod, and a light intensity of about 60 μmol m-2s-1.
-
Agrobacterium tumefaciens strain LBA4404 harboring the infectious clone of SPLCV-JS (Genbank accession No. FJ176701) was used for agroinoculation. The construction of this SPLCV-JS clone has been reported previously. In brief, a fragment containing ~1.3 units of the SPLCV genome was cloned into the pPZP100 binary vector to get the infectious clone pPZP-SPLCVJS (Bi H, et al., 2012). The DNA-A and DNA-β clones of Tomato yellow leaf curl China virus isolate Y10 (TYLCCNV-Y10), which have been described previously (Cui X F, et al., 2004), were generously provided by Professor Xueping Zhou, Zhejiang University, China.
-
A. tumefaciens harboring the infectious clone of SPLCV-JS was grown overnight at 28℃ in 300 mL YEB medium supplemented with 25 μg/mL rifampicin, 100 μg/mL streptomycin and 25 μg/mL chloromycetin. The cells were harvested by centrifugation and resuspended at a final concentration with an optical density (OD600)of 1.5-2.0 in a solution containing MS basal medium, 20 g/L sucrose, 10 mmol/L 2-(N-morpholino) ethanesulfonic acid (MES), 200 μmol/L acetosyringone, 0.005% Tween-20 and 0.04% Silwet L-77, pH 5.6. This inoculum was poured into a container e.g. a glass beaker), and sweet potato plantlets with 4-5 leaves were immersed in it. The beaker with the plants was placed in a vacuum chamber, which was then evacuated down to a pressure of -0.07 MPa for 10-15 min. After incubation, the vacuum was rapidly released, the plants were removed from the chamber and returned to the humidor to allow infiltration of the leaves with the bacterial suspension. The infiltrated plantlets were washed once in double-distilled water to remove the remaining infection solution. For single infection with SPLCV-JS, plants were agroinfiltrated with SPLCV-JS suspension in the infiltration medium. For SPLCV-JS or TYLCCNV-Y10 DNA-A infection concomitant with TYLCCNV-Y10 DNA-β, equal volumes of SPLCV-JS or TYLCCNV-Y10 DNA-A and TYLCCNV-Y10 DNA-β suspension were mixed together and co-infiltrated. A. tumefaciens harboring an empty vector was used as a negative control in these experiments. The vacuum infiltration experiment was repeated twice independently.
-
The symptoms of agroinfected plants were evaluated weekly by recording the leaf phenotype.
-
Total genomic DNA was extracted from the young upper leaves of agroinfected plants at 35 days postinoculation (dpi) according to the method of Soni and Murray (Soni R, et al., 1994). The genomic DNA was checked for the presence of SPLCV-JS (C1-5FP/C1-3RP) or TYLCCNV-Y10 DNA-β (beta01/beta02; Briddon R W, et al., 2002) by PCR assay using the primer pairs C1-5FP(5'-AGGGAGCTAAATCCTCGTCC-3')/C1-3RP (5'-TCAGGAACTTTCCTCTTGGC-3') or beta01 (5'-GGTACCACTACGCTACGCAGCAGCC-3')/beta02 (5'-GGTACCTACCCTCCCAGGGGTACAC-3'), respectively. PCR assays were conducted in a reaction volume of 50 μL containing 50 ng DNA template, 0.4 μmol/ L forward primer, 0.4 μmol/L reverse primer, 10 mmol/L Tris-HCl (pH 8.5), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.2 mmol/L of each dNTP and 1 U Taq DNA polymerase. The PCR assay was performed with an initial denaturation for 5 min at 95℃, followed by 30 cycles at 95℃ for 40 sec, 56℃ for 1 min, 72℃ for 1 min (for C1 gene) or 1.5 min (for betasatellite) and a final extension at 72℃ for 10 min. The predicted lengths of the amplified fragment of C1 and betasatellite were 821 bp and 1, 356 bp, respectively. The genomic DNA of field-collected sweet potato Xu 35-5 plants infected by SPLCVJS, and the Xushu 22 plants vacuum-infiltrated with the TYLCCNV-Y10 DNA-A and DNA-β suspension were used as the positive controls.
For Southern blot analysis, an aliquot of 10 μg of total DNA from each sample was digested with HindⅢ and analyzed using a standard protocol (Sambrook J, et al., 2001). Amplified viral DNA was hybridized with a digoxigenin (DIG)-labeled probe specific to the entire SPLCV-JS genome. Labeling, hybridization and chemiluminescent detection were performed according to the instructions of the manufacturer (Roche Applied Science, Germany). Field-collected sweet potato Xu 35-5 plants infected by SPLCV-JS were used as the positive controls.
Plant materials
Virus isolate and bacterial strain
Vacuum infiltration
Symptom evaluation of infected plants
Virus detection in infected plants by PCR and Southern blot analysis
-
Sweet potato plantlets with 3-4 leaves were agroinoculated with the infectious SPLCV-JS clone by vacuum infiltration. Details of the protocol for vacuum infiltration with the SPLCV-JS clone are shown in Figure 1. At 20 dpi, all the infected sweet potato plants remained asymptomatic. At a late infection stage (35 dpi), mild symptoms (yellowing) were observed in newly emerged leaves (Figure 2B, arrows); mock-inoculated plants did not show any symptoms (Figure 2A). Symptom evaluation, together with PCR detection, identified five infection-positive plants among the 20 that were inoculated with SPLCV-JS (Figure 3A; Table 1). Only symptomatic plants were PCR-positive. In addition, Southern blot analysis using a probe specific for the SPLCV-JS genome revealed the presence of the typical viral DNA forms, i.e., single-stranded genomic DNA (ssDNA) and double-stranded replicative DNA (Lin) of begomovirus, in agroinoculated Xushu 22 sweet potato plants (Figure 3B, lanes 2 and 3). The experiment was repeated twice independently with similar results. These data support the successful infection of the sweet potato cultivar Xushu 22 by the infectious SPLCV-JS clone using vacuum infiltration.
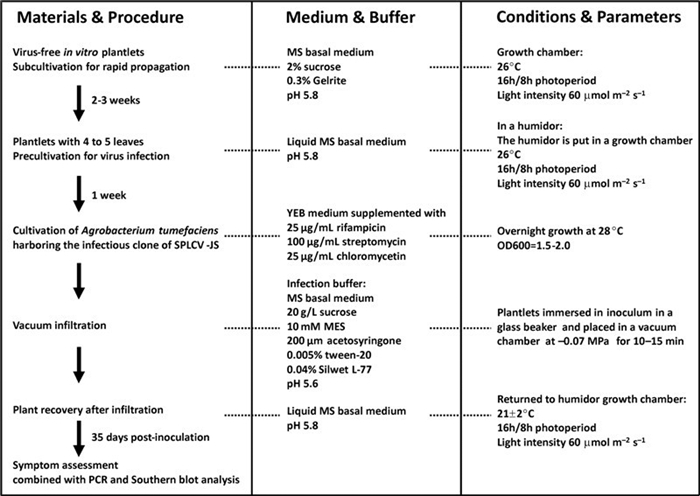
Figure 1. Protocol for vacuum agroinfiltration of sweet potato cultivar Xushu 22 with the SPLCV-JS infectious clone.
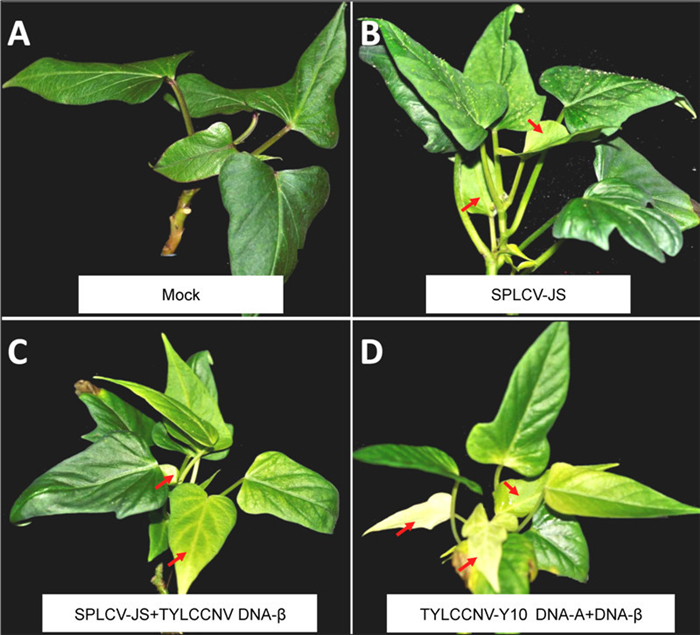
Figure 2. Symptoms of sweet potato cultivar Xushu 22 infected with the SPLCV-JS infectious clone by vacuum agroinfilitration. A, Mock control; B, SPLCV-JS; C, SPLCV-JS and TYLCCNV-Y10 DNA-β; and D, TYLCCNV-Y10 DNA-A and DNA β. Symptoms in the leaves are indicated by red arrows.

Figure 3. SPLCV viral DNA accumulation in sweet potato plants infected with the SPLCV-JS infectious clone as determined by PCR detection and Southern blot analysis. Panel A, PCR amplification of the SPLCV-JS C1 gene; Panel B, Southern blot analysis of SPLCV accumulation using a DIG-labeled probe specific to the entire SPLCVJS genome. Geminiviral DNA forms are marked as open circular double-stranded (OC), linear double-stranded (Lin), supercoiled double-stranded (SC), or single-stranded (SS). Panel C, PCR detection of TYLCCNV-Y10 DNA-β. M, Molecular marker; Lane 1, Mock infected (MI); Lanes 2 and 3, SPLCV-JS; Lanes 4 and 5, SPLCV-JS and TYLCCNV-Y10 DNA-β; Lane 6, Field-collected sweet potato Xu 35-5 infected by SPLCV-JS as positive control in panels A and B or the Xushu 22 plants infected by TYLCCNV-Y10 DNA-A and DNA-β as positive controls in panel C.
Inoculum Infected/inoculated plants (n) Symptoms SPLCV-JS 5/20 Mild yellowing in newly emerged leaves SPLCV-JS + TYLCCNV-Y10 DNA-β 6/20 Obvious leaf yellowing and vein clearing TYLCCNV-Y10 DNA-A + DNA-β 2/5 Severe leaf yellowing Empty vector 0/5 Symptomless Empty vector + TYLCCNV-Y10 DNA-β 0/5 Symptomless Table 1. Infectivity analysis of the SPLCV-JS infectious clone in sweet potato cultivar Xushu 22 plants.
-
Our previous study reported a synergistic effect of that SPLCV-JS and a heterologous betasatellite DNA(TYLCCNV-Y10 DNA-β)on enhanced symptom severity and viral DNA accumulation in N. benthamiana (Bi H, et al., 2012). We further tested that effect in sweet potato plants agroinoculated concomitantly with both SPLCVJS and TYLCCNV-Y10 DNA-β infectious clones. The enhancement of symptom severity by TYLCCNV-Y10 DNA-β was shown at 35 dpi by increased severity of obvious leaf yellowing and vein clearing in six of the 20 plants that were inoculated(Figure 2C, arrows). In addition, two of the five plants agroinoculated with TYLCCNV-Y10 DNA-A and DNA-β developed severe leaf yellowing in the new leaves, somewhat similar the appearance of an albino seedling(Figure 2D, the arrows). The experiment was repeated twice independently with similar results.
In Southern blot analysis, the accumulation of replicative DNA forms including open circular double-stranded (OC), supercoiled double-stranded (SC), and single-stranded (ss) DNA were observed in the plants inoculated with SPLCV-JS and TYLCCNV-Y10 DNA-β (Figure 3B, lanes 4 and 5). The accumulation of SPLCVJS viral DNA was more obvious in these mix-infected plants than that seen with SPLCV-JS infection alone. No signal was detected in the mock-infected control plants (Figure 3B, lane 1) using a probe specific for the entire SPLCV-JS genome. Additionally, a field-collected, diseased Xu 35-5 s weet potato sample plant that was used to amplify the SPLCV-JS genome and served as a positive control showed a potent SPLCV signal (Figure 3B, lane 6). PCR assays confirmed the presence of DNA-β in sweet potato plants coinoculated with SPLCV-JS and TYLCCNV-Y10 DNA-β (Figure 3C, lanes 4-5) and in plants infected with TYLCCNV-Y10 DNA-A and DNA-β (Figure 3C, lane 6). Thus, we showed that SPLCV-JS interacts synergistically with the heterologous betasatellite in sweet potato, much like the results seen in N. benthamiana (Bi H, et al., 2012).
An infectious SPLCV-JS clone was delivered into sweet potato cultivar Xushu 22 by vacuum infiltration
The symptom severity of SPLCV-JS in sweet potato culitivar Xushu 22 could be enhanced by a heterologous betasatellite DNA
-
Construction of infectious virus clones is an essential molecular tool for studying viral biological properties, validating the function of viral genes, and screening virus-resistant germplasm. The agroinoculation of viral clones using Agrobacterium and their binary vectors is a cost-effective, highly efficient method for analyzing the infectivity of plant viruses, and is widely used by geminivirologists (Boulton M I, 1996; Carrillo-Tripp J, et al., 2006; Elmer J S, et al., 1988; Grimsley N, et al., 1987).
However, several attempts to achieve infection using cloned sweepoviruses were unsuccessful (Lotrakul P, et al., 2003; Paprotka T, et al., 2010). Recently, successful infection of N. benthamiana and several species of Ipomoea by a clone of sweet potato leaf curl Lanzarote virus (SPLCLaV), an infectious sweepovirus from Spain, was reported using syringe infiltration (Trenado H P, et al., 2011). Our previous study demonstrated that the sweepovirus clone SPLCV-JS from China was infectious in N. benthamiana (Bi H, et al., 2012). In this work, we describe a protocol for agroinoculation of the sweet potato cultivar Xushu 22, which is widely grown in China, with the infectious sweepovirus clone SPLCV-JS using vacuum infiltration.
Most previous studies of SPLCV infectivity have been based on transmission by the whitefly Bemisia tabaci (Ling K S, et al., 2010; Simmons A M, et al., 2009, Valverde R A, et al., 2004). However, the transmission rate of sweepoviruses to sweet potato plants by whiteflies is very low, and it has been suggested that such low transmission rates may reflect the low amino acid sequence identity between the coat protein of sweepoviruses and those of other begomoviruses (Valverde R A, et al., 2004). Sweet potato mild mottle virus (SPMMV), and sweet potato chlorotic stunt virus (SPCSV), can also be transmitted by B. tabaci. This can lead to mixed infe ction because whiteflies transmit SPCSV more efficiently than SPLCV (Valverde R A, et al., 2004). Compared with whitefly transmission, agroinoculation with an infectious clone is more effective, and this has been a major challenge in the study of sweepoviruses (Trenado H P, et al., 2011). The development of an agroinoculation system with SPLCV-JS using vacuum infiltration in sweet potatoes is thus important for studying the virus in its endemic regions.
The symptoms caused by the SPLCV-JS infectious clone in sweet potato Xushu 22 were mild (Figure 2B), and the accumulation of this virus was relatively low (Figure 3B). In fact, asymptomatic begomovirus infections with mild symptoms are not uncommon in sweet potato plants (Clark C A, et al., 2006; Valverde R A, et al., 2007). However, field-grown sweet potato plants are often infected by multiple viruses (Valverde R A, et al., 2007), and it is highly possible that other viruses interact synergistically with sweepoviruses, leading to enhanced viral accumulation and symptom severity. Because an infectious SPLCV-JS clone is now available, direct cause-and-effect evaluation of symptoms or loss in yields associated with sweepovirus infection in sweet potato plants can be studied and demonstrated experimentally. The severity of symptoms and virus accumulation of SPLCV-JS were clearly enhanced in plants coinfected with SPLCV-JS and TYLCCNV-Y10 DNA-β. This observation indicated that in sweet potato Xushu 22, SPLCV-JS could interact synergistically with this heterologous betasatellite.
Since a virus-induced gene silencing (VIGS) system based on TYLCCNV-Y10 DNA-β has already been developed in N. benthamiana and tomato plants (Tao X R, et al., 2004), the interaction between SPLCV-JS and TYLCCNV-Y10 DNA-β provides a possibility for creating a VIGS system for gene function analysis and mechanistic investigations in the sweet potato.
-
This work was supported by grants from the National High Technology Research and Development Program of China (2012AA101204), Shanghai Municipal Afforestation and City Appearance and Environmental Sanitation Administration (G102410, F132427).
-
All the authors declare that they have no competing interests. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
Huiping Bi participated in the design of the study, performed the experiments, and drafted the manuscript. Peng Zhang conceived and designed the study and wrote the manuscript. All authors have read and approved the final manuscript.













 DownLoad:
DownLoad: