-
Dear Editor,
In the late stages of the retroviral life cycle, the transcription of viral pre-mRNA is initiated at the 5′ long terminal repeat (5′ LTR) and terminated at the 3′ LTR; the full-length and spliced viral RNAs are transported out of the nucleus and serve as templates for the translation of viral proteins or alternatively as the full-length viral genome to be packaged into virus particles. Antisense transcription has been described in the Retroviridae family, including in human T cell leukemia virus (HTLV), human immunodefciency virus (HIV), and feline immunodefciency virus (FIV) (Briquet et al., 2001; Cavanagh et al., 2006; Landry et al., 2007). Unlike delta-retroviruses such as HTLV, HIV and FIV belong to the Lentivirus genus. The antisense transcript of HTLV-1 has been intensively studied. It initiates at various positions in the 3′ LTR and encodes a functional protein, termed HBZ, for HTLV-1 bZIP factor (Cavanagh et al., 2006; Matsuoka and Green, 2009). HBZ negatively regulates basal and Tax-dependent HTLV-1 transcription via interactions between its bZIP (basic leucine zipper) domain and several transcription factors. It also promotes T-lymphocyte proliferation (Arnold et al., 2008) and induce T-cell lymphoma and systemic inflammation (Satou et al., 2011). The antisense transcript of HIV-1 is transcribed via the promoter located in the 3′ LTR and encodes a conserved antisense protein located between the surface (SU) and transmembrane (TM) proteins (Landry et al., 2007; Torresilla et al., 2013). In a previous report, a conserved open reading frame (ORF) in the antisense transcripts of FIV was predicted and antisense transcription was eventually demonstrated (Briquet et al., 2001). Antisense transcription has also been confrmed in murine leukemia virus (MLV), which is classifed as a gamma-retrovirus (Rasmussen et al., 2010).
However, there are limited data regarding antisense transcripts of retrovirus other than HTLV, HIV, FIV, and MLV. Thus, it is unknown whether antisense transcription is a universal phenomenon in retroviruses. In this study, we attempted to detect antisense transcription in bovine immunodeficiency virus (BIV). We predicted potential transcription start sites and polyA signal sites along the antisense proviral sequence of BIV127, a well-described BIV clone of the R29 strain (Garvey et al., 1990), using the Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/) and DNA PolyA Signal Miner (http://dnafsminer.bic.nus.edu.sg/). A potential polyA signal site was found at the site of nt 5800 (all positions are indicated with respect to NCBI RefSeq No. NC_001413.1). Upstream of this polyA site, two potential transcription start sites were predicted, located at the 3′ LTR and the TM protein-coding region. A 408-bp ORF (from nt 6934 to 6527) was found between the predicted transcription start sites and the potential polyA site, overlapping with the region encoding the SU protein (Figure 1A). The predicted protein encoded by this ORF was 136 amino acids in length and had an estimated molecular weight of 16 kDa.
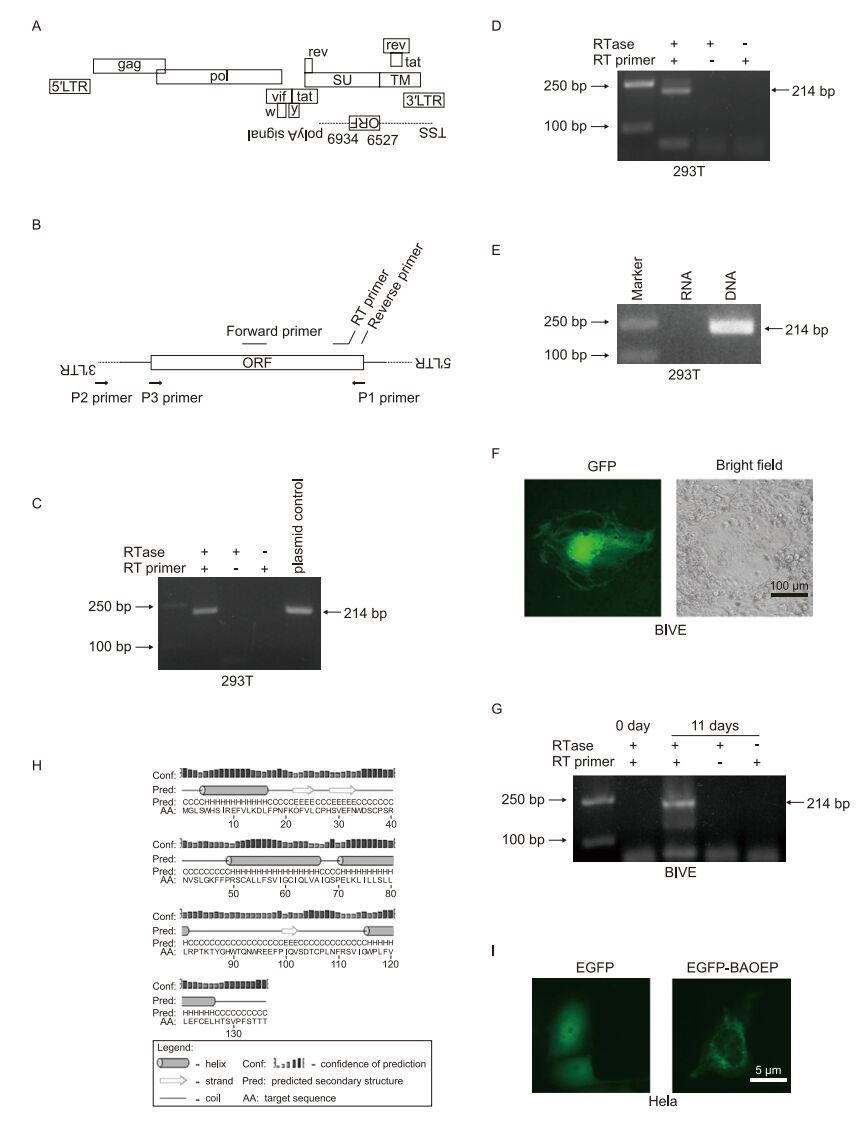
Figure 1. Detection of antisense transcripts in bovine immunodefciency virus (BIV). (A) Schematic diagram of the antisense open reading frame (ORF) in the BIV genome. (B) Primers used in this study. The sequences of these primers are listed in Table 1. (C) RT-PCR analyses of antisense transcripts on RNA samples from pBIV127-transfected 293T cells. (D) A region from the predicted ORF to the 3′ LTR was amplifed by PCR using primers P1 and P2, and the PCR products were transfected into 293T cells. The antisense transcript was detected using RT-PCR. (E) PCR products using primers P1 and P3 were transfected into 293T cells. RNA was extracted for RT PCR detection of the antisense transcript. Total DNA was extracted and used as a template for PCR with forward. Syncynia formation and reverse primers to serve as a positive control. (F) BIVE cells were infected with the BIV127 virus for 11 days and GFP expression were observed. (G) RT-PCR analyses for antisense transcripts on RNA samples from BIVE cells infected with BIV127 for 0 day and 11 days. (H) Prediction of secondary structures for the BIV antisense ORF-encoded protein (BAOEP) using PSIPRED v3.3 (http://bioinf.cs.ucl.ac.uk/psipred/). (I) Subcellular distribution of the EGFP-tagged BAOEP in HeLa cells.
We used strand-specific reverse transcription-polymerase chain reaction (RT-PCR) (Craggs et al., 2001) to detect the antisense transcript. To ensure high specifcity and exclude false-positive amplification from plasmid DNA or positive-strand viral RNA, the following steps were taken: 1. Tagged RT primer was used to increase specificity; 2. RNA samples were treated with DNase I to eliminate the contaminated plasmid DNA before reverse transcription; and 3. Two negative controls without the addition of RTase or RT primer were used. The tagged RT primer and forward primer were located in the predicted ORF, as shown in Figure 1B. The primer sequences used in this article are presented in Table 1. The proviral pBIV127 plasmid was transfected into 293T cells, and total RNA was extracted 24 h post transfection. RNA samples were treated with DNase I (Takara, Otsu, Japan). Then, RT reactions were performed with 1 μg of RNA and 1 μmol/L RT primer using Reverse Transcriptase M-MLV (Takara) according to the manufacturers' instructions. For the RTase-/RT primer+ control, 50% glycerol was added instead of reverse transcriptase; for the RTase+/RT primer-control, RT primer was not added to the reverse transcription reaction, but was added after the reaction with a final concentration of 1 μmol/L. cDNA samples were diluted 1, 000-fold and 2 μL of the diluted cDNA was used for the PCR reaction to eliminate amplification of segments on the wrong strand (Craggs et al., 2001). PCR was performed in a 25-μL volume with 0.1 μmol/L forward and reverse primer using PrimeSTAR HS DNA Polymerase (Takara). PCR products were analyzed by 2% agarose gel electrophoresis. In our experiments, excess RT primer in the cDNA was not removed before the PCR reaction. Though the RT primer had the potential ability to result in specific amplification using plasmid DNA or cDNA derived from positive-strand viral RNA (Craggs et al., 2001), the occurrence of false-positives was ruled out using the controls. For the control without the addition of reverse transcriptase during the cDNA synthesis reaction, plasmid contamination resulting in amplifcation would lead to a 214-bp band. Alternatively, cDNAs from the positive strand might be generated by auto-priming events during reverse transcription, and residual RT primer in the PCR reaction might enable these cDNAs to be amplifed. However, another control in which RT primer was not added during the cDNA synthesis reaction, but was added to the reaction products, would exclude this possibility. A band with the predicted length of 214 bp was detected in the reaction with the addition of RTase and RT primer, but not in the two negative controls (Figure 1C). This demonstrated the existence of antisense transcripts in 273T cells after pBIV127 transfection.

Table 1. Primers used in the study.
Though transfection of pBIV127 generated antisense transcripts, transcription might start at the plasmid backbone instead of the BIV genomic region. To address this issue, a segment of the BIV pre-genome from the predicted ORF to the 3′ LTR was amplifed by PCR, using primers P1 and P2, as illustrated in Figure 1B. The PCR products using primers P1 and P2 were transfected into 293T cells and antisense transcripts were detected (Figure 1D). For the negative control, PCR amplification was performed using primers P1 and P3. The PCR products were transfected into 293T cells and antisense transcripts were not detected (Figure 1E, lane labeled "RNA"); a specific band could be amplified from DNA extracted from the transfected cells, indicating successful transfection (Figure 1E, lane labeled "DNA"). These results demonstrated that antisense transcripts could be generated in the BIV pre-genome segment lacking the plasmid backbone sequence.
Next, we attempted to detect BIV antisense transcripts in BIV infected cells. BIV viruses were packaged in 293T cells by transfection of pBIV127 and then 293T cells were lysed by repeated cycles of freezing and thawing. Cell lysates were used to infect BIVE cells, which were from a BIV-permissive cell line with a GFP marker under the control of the BIV LTR to indicate BIV infection (Yao et al., 2010). After infection for 4 h, 293T cell lysates were removed, and adherent BIVE cells were washed with phosphate-buffered saline. Syncytia formation and GFP expression were observed at 11 days after infection, indicating successful infection by BIV (Figure 1F). RT-PCR was performed and antisense transcripts were detected in BIV-infected BIVE cells for the sample of BIVE cells infected by BIV for 11 days (Figure 1G).
The secondary structure of the BIV antisense ORF-encoded protein (BAOEP) was predicted using PSIPRED v3.3 (http://bioinf.cs.ucl.ac.uk/psipred/). Four helix regions were predicted, as shown in Figure 1H. We cloned the antisense ORF into the pEGFP-C1 vector to express EGFP-tagged BAOEP. The pEGFP-C1 vector expressing EGFP was used as a control. The distribution of EGFP-tagged BAOEP in HeLa cells was observed. EGFP-tagged BAOEP was mainly observed in the cytoplasm and exhibited a perinuclear distribution with a filamentous and punctate fluorescent signal, while the EGFP control was evenly distributed throughout the cell (Figure 1I). A previous report showed that the antisense protein of HIV-1 co-localizes with LC3B, which is an autophagosome marker, in a punctate manner (Torresilla et al., 2013). However, we did not detect the co-localization of EGFP-tagged BAOEP with mCherry-tagged LC3B in HeLa cells (data not shown). Moreover, the expression of EGFP-tagged BAOEP in HeLa cells did not induce autophagy, based on the quantification of the LC3-Ⅱ protein expression level (data not shown). Thus, the antisense transcript of BIV might not be related to autophagy. In summary, this report documents the generation of antisense transcripts from clone127 of the R29 strain of BIV. An antisense ORF was predicted, and antisense transcripts were detected in 293T cells transfected with DNA containing the BIV127 proviral sequence. More importantly, the antisense transcript was detected in BIVE cells infected with BIV127. It should be noted that this research attempted to demonstrate the presence of antisense transcripts in BIV-127 of the R29 strain; thus, the results do not address whether the antisense transcripts exist in other BIV strains. Further investigations are needed to address this issue. Recently, an infectious clone of bovine foamy virus, a retrovirus which could infect cattle, was obtained (Bing et al., 2014). It would facilitate the research regarding antisense transcription in foamyvirus. These studies would advance our understanding of antisense transcription and contribute to the characterization of retroviruses.
HTML
-
We greatly appreciate the gift of pBIV127 from Dr. Charles Wood (University of Nebraska, Lincoln) and BIVE cells from Wentao Qiao (Nankai University). This study was supported by National Natural Science Foundation of China (81371820).The authors declare that they have no competing interests. This article does not contain any studies with human or animals subjects performed by any of the authors.














 DownLoad:
DownLoad: