HTML
-
Novel human pathogens continue to emerge, the majority of which originate in wildlife (Jones et al., 2008). Bats are the only flying mammals. They make up approximately 20% of extant mammalian diversity, representing the second largest order of mammals after rodents. Bats are found on all continents, except in polar regions and a few oceanic islands. The diversity of bat species and some of their unique biological features have allowed them to serve as natural reservoir hosts for many viruses (Shi, 2013). Some bat species carry emerging or reemerging infectious disease agents, including severe acute respiratory syndrome coronaviruses (Lau et al., 2005; Li et al., 2005; Ge et al., 2013), henipaviruses (Halpin et al., 2000; Chua et al., 2002), and Ebola viruses (Pourrut et al., 2009). These viruses are pathogenic to humans and other mammals, but appear to cause no clinical disease in bats during natural or experimental infection (Williamson et al., 1998; Williamson et al., 2000; Middleton et al., 2007). The long co-evolutionary history of bats and viruses may have resulted in the adaptation of bats' immune systems to cope with viral infection. It is hypothesized that bat innate immunity allows bats to limit viral replication to very low levels, allowing persistent infections of viruses in bat populations (Papenfuss et al., 2012). However, because little information is currently available on any aspect of bat immunology and few bat-specific reagents exist, this hypothesis remains untested.
One of the earliest immune responses initiated after viral infection is the production of interferons (IFNs). IFNs provide the first line of defense against viral infection and play a pivotal role in complex cytokine networks (Randall and Goodbourn, 2008). Type Ⅰ IFNs, some of the most important antiviral immune factors, are associated with the host's ability to reduce viral replication by inducing the expression of IFN-stimulated genes (ISGs).
To underst and bat-virus interactions, the Australian black flying fox, Pteropus alecto, has been used as a model bat species (Crameri et al., 2009). This species is the reservoir of a variety of closely related viruses, including rabies virus, Hendra virus, and Nipah virus (Marsh and Wang, 2012). Some genes involved in the immune responses of P. alecto have been identified, and functional studies of these genes have been performed (Baker et al., 2010; Cowled et al., 2011; Virtue et al., 2011; Zhou et al., 2011a; Zhou et al., 2011b; Cowled et al., 2012). Type Ⅰ IFNs were recently described in two other species of fruit bats, the Egyptian rousette, Rousettus aegyptiacus; the Malaysian flying fox, Pteropus vampyrus (Omatsu et al., 2008; Kepler et al., 2010); and two species of microbats, Rhinolophus affinis and Rhinolophus sinicus (Li et al., 2015). In this study, we analyzed the sequence and antiviral activity of IFNβ cloned from a Chinese microbat species, Myotis davidii. Our results demonstrate that IFN receptor (IFNR) binding site sequences of IFNβ are conserved and that in vitro antiviral activity is similar across many mammalian genera, including Myotis.
-
Culture conditions for bat cell lines have been described previously (Li et al., 2010). The cell lines used in this study included an M. davidii primary kidney cell line (MdKi, previously called BK) and an immortalized Rhinolophus sinicus kidney cell line (RsKT). The MdKi cells were cultured with RPMI 1640 medium, supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. The RsKT cells were cultured in DMEM/F12-Hams medium, supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. All cells were maintained at 37 ℃ in an incubator supplemented with 5% CO2.
-
Bat adenovirus TJM (BtAdV-TJM) was grown in MdKi cells for 48 h in a humidified atmosphere of 5% CO2 at 37 ℃ (Li et al., 2010). The virus-containing supernatant was collected and the 50% tissue culture infective dose (TCID50) was determined. Vesicular stomatitis virus-green fluorescent protein (VSV-GFP) (Fernandez, 2002) was grown in 293T cells for 48 h, the virus-containing supernatant was collected, and the TCID50 was determined. Sendai virus (SeV) was propagated in 10-day-old embryonated eggs, and the viral titer was determined by hemagglutination assays using chicken red blood cells.
-
MdKi cells were seeded at 1×106 cells/well in 6-well tissue culture plates. The cells were stimulated by transfection with 2 μg/mL polyI:C (InvivoGen) using 5 µL of Lipofectamine 2000 (InvivoGen) or by infection with SeV at 100 hemagglutinating units (HAU) /well. After stimulation, the cells were cultured in a humidified atmosphere of 5% CO2 at 37 ℃. The cells were harvested in RLT buffer (Qiagen) at 0, 6, 12, and 18 h post-infection (hpi) and stored at −80 ℃ until the RNA was extracted.
-
The complete IFNB gene was identified in the M. davidii genome (GenBank accession no: XM_006764946). PCR and random amplification of cDNA ends (RACE) primers (Table 1) were designed based on the M. davidii genomic sequence. MdKi cells were infected with SeV and total RNA was extracted 6 hpi for use as a template for IFNB gene amplification. The samples were homogenized using QIAshredder (Qiagen). Total RNA was extracted using the RNeasy Mini Kit (Qiagen) with on-column DNase Ⅰ treatment (Qiagen) to remove all traces of genomic DNA. The full-length sequence of IFNB was obtained using 5′ and 3′ RACE with the Full RACE Kit (Takara). The DNA amplification conditions included an initial denaturation step at 95 ℃ for 5 min; 40 cycles of denaturation at 94 ℃ for 30 s, annealing at 53 ℃ for 30 s, and extension at 72 ℃ for 50 s; and a final extension at 72 ℃ for 10 min.
-
The PCR and RACE products were cloned into the pGEM-T Easy Vector (Promega) for sequencing. The GenBank accession numbers of the sequences used in the phylogenetic analysis are as follows: NP_001092910, Equus caballus (horse); NP_002167, Homo sapiens (human); XP_528553, Pan troglodytes (chimpanzee); NP_034640, Mus musculus (mouse); NP_062000, Rattus norvegicus (rat); NP_001129259, Canis lupus familiaris (dog); AAA31056, Sus scrofa (pig); NP_776775, Bos taurus (cattle); XP_006765009, Myotis davidii; XP_006098187, Myotis lucifugus; ELK06976, P. alecto; BAF37103, Rousettus aegyptiacus; NP_001020007, Gallus gallus (chicken); XP_002708014, Oryctolagus cuniculus (rabbit); KM056305, Rhinolophus sinicus (Chinese rufous horseshoe bat); and KM056306, Rhinolophus affinis (intermediate horseshoe bat). All sequence alignments were performed with the ClustalW program and visualized with GeneDoc software (Thompson et al., 1994). Signal peptides were identified with SignalP version 3.0. Protein domains were identified using the NCBI Conserved Domains search tool (Marchler-Bauer et al., 2011). The phylogenetic tree was generated in MEGA 6 using the neighbor-joining method with 1000 bootstrap replicates.
-
The bat IFNB was cloned into the pET32a expression vector and was used to transform Escherichia coli strain DH10B. The clones were analyzed with restriction digestion to confirm the correct orientation of IFNB. Briefly, Escherichia coli strain BL21 (pET32–IFNβ) was cultured, induced with isopropyl-β-D-thiogalactoside, and then harvested by centrifugation. The sonicated cell lysates were treated with Triton X-100. The proteins were then immobilized on a metal affinity chromatography column and eluted with imidazole. Samples for analysis were collected at each purification step and separated on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel. Western blotting was performed with a His-tag-directed antibody as the primary antibody (1:1000) and an alkaline-phosphatase-conjugated goat anti-rabbit IgG antibody (1:2000, Sino-American) as the secondary antibody.
-
A VSV–GFP bioassay was performed to determine the antiviral activity of the expressed recombinant bat IFNβ. MdKi cells and RsKT cells were seeded in 96-well plates at a density of 1×104 cells/well. After 24 h, the bat cells were left untreated or were treated for 24 h with five concentrations of bat IFNβ diluted in medium and then incubated with VSV–GFP at a multiplicity of infection of 1. After adsorption for 1.5 h at 37 ℃ in a 5% CO2 incubator, the inoculum was removed and 100 μL of medium was added to each well. GFP expression in the cells was visualized by fluorescence microscopy at 18 hpi. The number of fluorescent points was counted to evaluate the inhibitory effect of bat IFNβ.
-
Similar assays were performed in MdKi and RsKT cells using the bat adenovirus, BtAdV. The two bat cell lines were inoculated with BtAdV at a multiplicity of infection of 1 and incubated for 1 h at 37 ℃. The virus-containing medium was then replaced with culture medium, and the cultures were incubated for 24 h in a humidified atmosphere of 5% CO2 at 37 ℃. The culture supernatant was collected and the TCID50 was determined by observation of the cytopathic effect.
-
Quantitative Real-time PCR (qRT-PCR) was performed on the total RNA extracted from the cells. In brief, total RNA was used for the synthesis of cDNA with the PrimeScript® RT Reagent Kit with gDNA Eraser and SYBR® Premix Ex TaqTM Ⅱ for real-time PCR (Takara). The relative gene expression after treatment was quantified with the StepOnePlusTM Real-Time PCR System (Applied Biosystems), and the comparative threshold cycle (Ct) method was used to calculate the fold change in gene expression. Relative gene expression was calculated using the mean values obtained from the arithmetic formula 2–∆∆Ct (Applied Biosystems). The cycling profile consisted of an initial denaturation at 95 ℃ for 30 s, and 40 cycles of 95 ℃ for 5 s and 60 ℃ for 30 s, followed by a melting curve analysis. All data were normalized to the housekeeping gene for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Cell lines
Viral culture
Stimulation of bat cells with poly I:C treatment and SeV infection
PCR amplification of the M. davidii IFNβ gene (IFNB)
Sequencing and phylogenetic analysis
Expression of bat IFNβ in Escherichia coli and purification of the protein
Antiviral activity assay of expressed recombinant bat IFNβ against VSV–GFP infection in bat cells
Antiviral activity assay of bat IFNβ against bat adenovirus in bat cells
Quantitative Real-time PCR
-
The expected DNA fragment of 561 bp was amplified from bat cells and sequenced. The deduced protein encoded by IFNB contained many features that are conserved in the IFNβ proteins of other species, including a signal-peptide cleavage site and several residues that are responsible for the binding of IFNβ to the type Ⅰ IFNR (IFNAR) (Figure 1A).
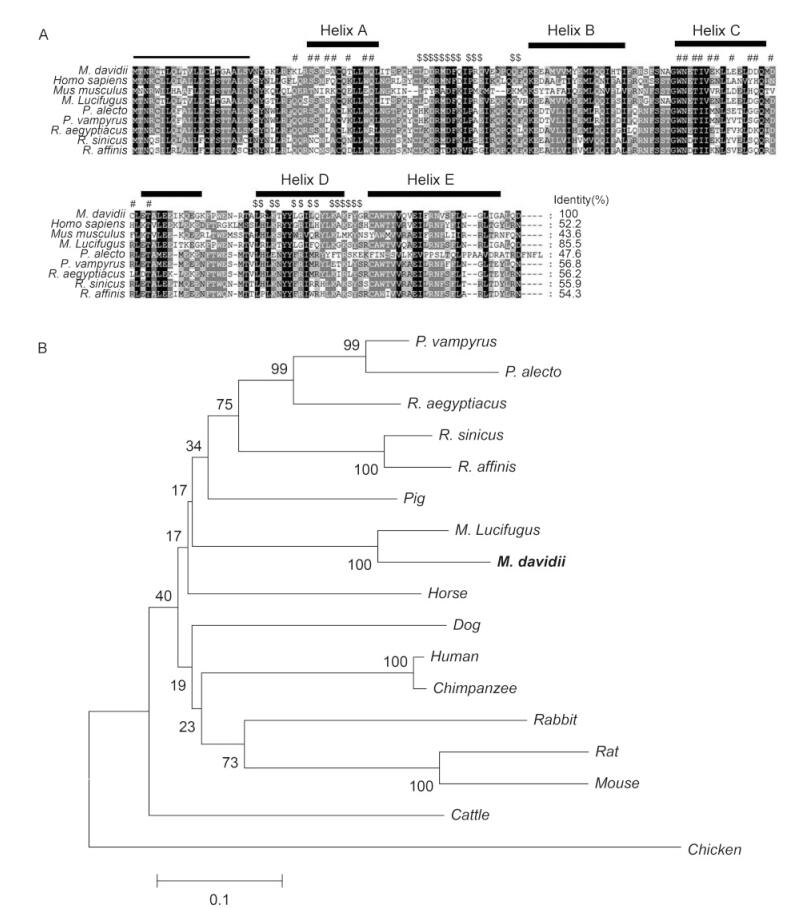
Figure 1. Sequence analysis of Myotis davidii IFNβ. (A) ClustalW-generated multiple amino acid sequence alignment of M. davidii and other bats, mice, and humans IFNβ. The predicted signal peptides are underlined. The putative IFNAR1-binding sites and IFNAR2-binding sites are indicated with "#" and "$", respectively. The secondary structure of IFNβ is shown above the alignment. Five helices, designated Helix A–E, are indicated with boxes. (B) Phylogenetic analysis based on an amino acid alignment of bat IFNβ with those of representative vertebrate species. Percentage branch support calculated with 1000 bootstrap replicates is indicated when it exceeds 60%. M. davidii IFNβ is highlighted in bold. The accession numbers of the IFNβ protein sequences used in this analysis are listed in the Materials and Methods.
Three-dimensional modeling of the bat IFNβ structure revealed five putative a-helices (A–E) (Arnold et al., 2006). The M. davidii IFNβ sequence shares 43%–85% amino acid identity with IFNβ proteins of other mammalian species, sharing the highest similarity with IFNβ of another Myotis species. A phylogenetic analysis of bat IFNβ and other IFNβ sequences from a variety of vertebrates was performed. As previously described (Zhang et al., 2013), M. davidii IFNβ clustered with M. lucifugus IFNβ within the laurasiatherian mammalian sequences (Figure 1B).
-
To examine the expression patterns of bat IFNs in response to polyI:C treatment or viral challenge, we used polyI:C and SeV to stimulate MdKi cells. The total RNA in the cells was isolated at different time points after infection. Successful infection by SeV was confirmed by RT-PCR (data not shown). The expression kinetics of IFNB and ISGs were determined by qRT-PCR. Following SeV infection, expressions of IFNB and ISGs were induced at 6 hpi and peaked at 12 hpi (Figure 2A). Similar to SeV infection, treatment with the double-str and ed RNA analog polyI:C also induced IFNB and ISG expression (Figure 2B).
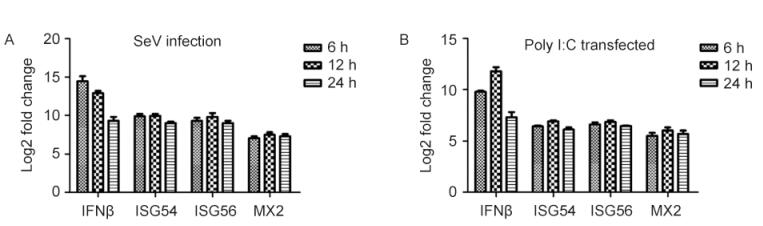
Figure 2. Analysis of bat IFNB and ISG expression in MdKi cells after treatment with polyI:C (A) or infection with SeV (B). Cells were transfected with polyI:C or infected with SeV and collected at the indicated time points. IFNB and ISG mRNAs were measured by qRT-PCR. The data were normalized to the housekeeping gene GAPDH. Data are the mean values of three separate experiments and the error bars represent standard errors.
-
The expressed recombinant IFNβ of the Myotis bat was analyzed with SDS-PAGE and confirmed with western blotting using an antibody directed against the His tag (Figure 3). To assess the antiviral activity of the recombinant bat IFNβ, MdKi and RsKT cells were pretreated with bat IFNβ. As shown in Figure 4, the treatment of MdKi and RsKT cells with IFNβ before VSV–GFP infection caused a significant dose-dependent reduction in the number of VSV–GFP-positive cells compared to those in the mock-treated cells. A similar inhibitory effect was observed during the infection of these two bat cell lines with BtAdV (Figure 5). These results demonstrate that recombinant bat IFNβ mitigated viral infection in bat cells in vitro.
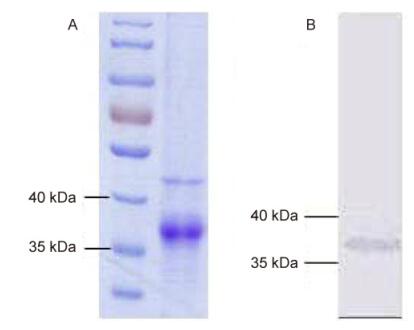
Figure 3. SDS-PAGE (A) and western blotting (B) analyses of recombinant M. davidii IFNβ expressed in Escherichia coli. (A) Purified recombinant M. davidii IFNβ protein was resolved with 12% SDS-PAGE and analyzed following Coomassie Brilliant Blue staining. (B) Purified recombinant M. davidii IFNβ was analyzed by western blotting, using an antiboday directed against the His tag.
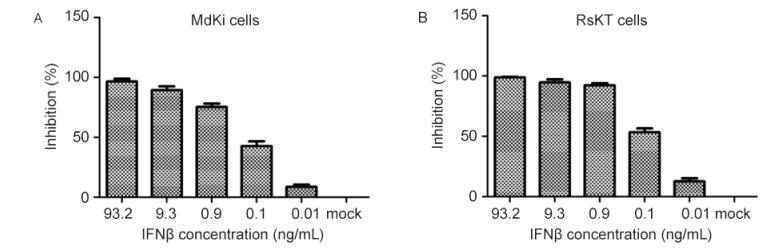
Figure 4. Antiviral activity assay of IFNβ against VSV–GFP in RsKT cells (A) and MdKi cells (B). Cells were treated for 24 h with five serial dilutions of IFNβ in medium and incubated with VSV–GFP. GFP expression in the cells was visualized and measured at 18 hpi.

Figure 5. Antiviral activity assay of bat IFNβ against bat adenovirus (BtAdV) in MdKi and RsKT cells. Cells were treated with serial dilutions of IFNβ or mock medium for 24 h and infected with BtAdV for another 24 h. The culture supernatant was collected and the TCID50 was determined by observation of the cytopathic effect.
Characterization of M. davidii IFNβ
Bat IFNs and ISGs were highly upregulated in bat cells after poly I:C treatment and SeV infection
Expressed recombinant bat IFNβ inhibited virus replication in bat cells
-
In this study, we cloned and characterized IFNβ from a Chinese microbat, M. davidii. Compared with IFNβ sequences from other mammals, the M. davidii IFNβ sequence shares highest amino acid identity with that of M. lucifugus (85%). A number of conserved features were identified in the bat IFNβ sequence, which is consistent with its functional equivalence to the IFNβ proteins of other species. To examine the expression pattern of bat IFNβ in response to polyI:C treatment or viral challenge, we used polyI:C and SeV to stimulate MdKi cells. The qRT-PCR results demonstrated that bat IFNB and ISGs were highly upregulated in the bat cells after polyI:C treatment or SeV infection. Furthermore, the expressed recombinant bat IFNβ protein exerted antiviral activity against human VSV and BtAdV. To our knowledge, this is the first study to report the antiviral activity of IFNβ in a microbat.
Previous studies have reported that recombinant type Ⅲ IFN and type Ⅱ IFN from the megabat P. alecto inhibited the replication of Pulau virus, Semliki Forest virus, and Hendra virus (Zhou et al., 2011a; Janardhana et al., 2012). Another research group characterized IFNκ and IFNω from the European serotine bat, Eptesicus serotinus. Both type Ⅰ IFNs have been shown to inhibit the replication of different lyssaviruses, to different extents, in susceptible bat cells (He et al., 2014). However, the antiviral activity of microbat IFNβ is less studied. Here, our results indicate that the M. davidii IFNβ protein has antiviral activity, consistent with type Ⅰ IFNs of other bat species. Our results also indicate that bat IFN has antiviral activity similar to that of other mammalian IFNs.
As in other mammals, the bat immune system comprises both innate and adaptive elements (Baker et al., 2013; Brook and Dobson, 2015). Recent studies have demonstrated that type Ⅰ IFNα and IFNβ from Rousettus aegyptiacus and type Ⅱ IFNγ and type Ⅲ IFNλ from P. alecto play crucial roles in virus interactions with these hosts (Zhou et al., 2011b; Janardhana et al., 2012). In addition to IFN sequence characterization, the induction of IFNs, their signaling pathways, and their antiviral activities following viral infection have also been investigated in cell lines derived from Eidolon helvum and P. alecto (Biesold et al., 2011; Virtue et al., 2011). These results provide evidence that the signaling molecules downstream of the IFN response in bats are probably similar to those in other mammals. In addition to the innate immune response, studies have examined the adaptive immune response in bats, demonstrating that bats do mount antibody responses to various viral infections, including lyssaviruses, filoviruses, and henipaviruses (Hayman et al., 2013). However, it seems that their humoral adaptive immunity is less effective at controlling viral transmission (Plowright et al., 2008; Turmelle et al., 2010; Brook, 2015). From the limited number of bat immune studies available, it seems that the bat has immunological features similar to those of other mammals.
Recently, the whole-genome sequences of two distantly related species, the fruit bat P. alecto and the insectivorous bat M. davidii, became available (Zhang et al., 2013). Comparative analyses indicated unexpected concentrations of positively selected genes in the DNA damage checkpoint and nuclear factor κB pathways, which might play a role in bat immunity. Because the DNA damage response plays an important role in host defenses and is a known target of virus interaction, a link between the metabolism and immunity of bats may explain the tolerance to viruses observed in bats (Wang et al., 2011; O'Shea et al., 2014). These issues warrant further investigation to determine whether these unique characteristics are associated with the asymptomatic nature of viral infections in bats.
-
This work was funded by the National Natural Science Foundation of China (No. 31321001).
-
All experiments were conducted under the guidelines of the Animal Ethics Committee in Wuhan Institute of Virology, Chinese Academy of Sciences. All the authors declare that they have no competing interest.
-
Y.L. and Z.S. conceived and designed the experiments; Y.L., L.W., and P.Z. performed the experiments; Y.L., L.W., Q.Z., and M.W. analyzed the data; X.Y. and X.G. provided the reagents and materials essential for the research; Y.L. wrote the paper.













 DownLoad:
DownLoad: