HTML
-
Coronaviruses(CoVs)are important infectious pathogens that are associated closely with respiratory and enteric diseases in humans and animals(Perlman and Netland, 2009; Belouzard et al., 2012; Li, 2013). CoVs have a single-strand positive sense RNA genome and consist of four groups: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus(Table 1).
Genus Species Receptor, co-receptor Alphacoronavirus HCoV-229E hAPN HCoV-NL63 ACE2 TGEV pAPN, Neu5Ac, Neu5Gc PEDV pAPN, Neu5Ac Betacoronavirus SARS-CoV ACE2 MHV CEACAM1 BCoV Neu5, 9Ac2 HCoV-OC43 Neu5, 9Ac2 Gammacoronavirus IBV Neu5Ac Deltacoronavirus Bulbul coronavirus HKU11 Unknown Thrush coronavirus HKU12 Munia coronavirus HKU13 Table 1. Coronavirus genera, species, and receptors.
Viral entry into cells is highly dependent on the interaction between viral particles and the host cells. CoVs use a variety of cellular receptors and co-receptors, including proteins and sugars, to facilitate their entry into cells. Infection begins with an interaction between the virus and its specific receptors(Li, 2015). Severe acute respiratory syndrome coronavirus(SARS-CoV)of group beta uses a type I integral membrane protein, angiotensin-converting enzyme 2(ACE2), as a receptor(Li et al., 2005). There is no structural homology between SARS-CoV and the Human coronavirus NL63(HCoV-NL63) receptor-binding domain(RBD), but they recognize the same receptor, ACE2(Wu et al., 2011). Although the RBD crystal structures of transmissible gastroenteritis virus(TGEV) and HCoV-NL63 are similar, they use different receptors(Reguera et al., 2012). Aminopeptidase N(APN), also known as CD13, is a type II transmembrane protein(Tusell et al., 2007). TGEV and porcine epidemic diarrhea virus(PEDV)infect cells through interaction with porcine(p)APN(Li et al., 2007; Nam and Lee, 2010). Human(h)APN is a receptor for HCoV-229E(Belouzard et al., 2012). In the same beta group, the receptors for mouse hepatitis virus(MHV) and bovine coronavirus(BCoV)are carcinoembryonic antigen-related cell adhesion molecule 1(CEACAM1) and a sugar, respectively, despite their high sequence homology(Peng et al., 2011; Peng et al., 2012). CEACAM1, the first identified coronavirus receptor(Dveksler et al., 1991), is a type I transmembrane multifunctional protein of the immunoglobulin superfamily(de Haan et al., 2005).
Cell entry and interspecies transmission of CoVs are mediated by the spike protein(S). CoV S is a class I fusion protein(Bosch et al., 2003). In CoVs, S is a significant surface protein and plays an important role in mediating infection of virions(Schwegmann-Wessels et al., 2009). S is also responsible for receptor binding and the fusion of viral and cellular membranes. All CoV S share the same functional component in two domains: an N-terminal domain called S1 that is responsible for receptor binding, and a C-terminal S2 domain that is responsible for fusion. S1 contains two subdomains, an N-terminal domain and a C-terminal domain(Figure 1A). Both function as RBDs and bind a variety of proteins and sugars(Belouzard et al., 2012).
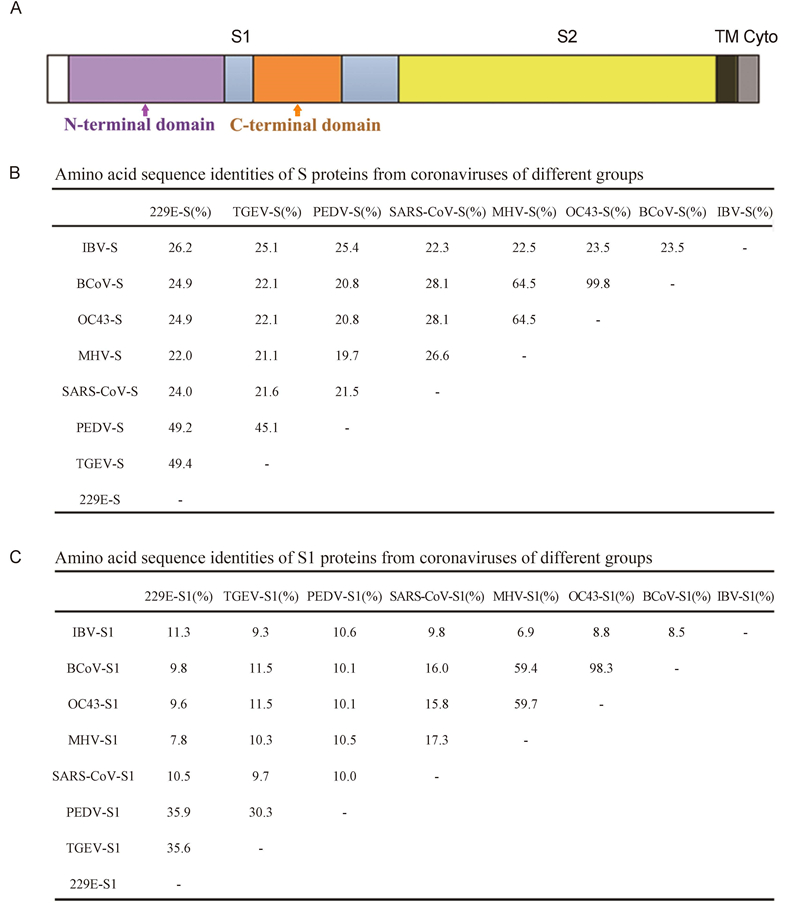
Figure 1. S proteins from coronaviruses of different groups. (A) Domain structure of the S protein, including the S1 and S2 regions, transmembrane domain (TM), and cytoplasmic tail (Cyto). (B) Amino acid sequence identities of S proteins from coronaviruses of different groups. The GenBank accession numbers are as follows: ABD72982.1 for SARS-CoV-S, AAR92025.1 for MHV-S, NP_073551.1 for HCoV-229E-S, ABG89335.1 for TGEV-S, NP_598310.1 for PEDV-S, AAT84362.1 for HCoV-OC43-S, ABM66810.1 for BCoV-S, and NP_040831.1 for IBV-S. (C) Amino acid sequence identities of S1 proteins from coronaviruses of different groups.
Because some CoVs have strong pathogenicity, the lentiviral pseudotype system is a reliable tool to study the proteins of highly pathogenic viruses under conventional biosafety conditions. HIV-Luc is an HIV-1 based lentiviral vector bearing the luciferase reporter gene and has been used in the production of pseudotyped viruses(Kang et al., 2012; Lu et al., 2012; Tang et al., 2012). Pseudovirus entry efficiency is characterized based on the luciferase activity. Viral particles pseudotyped by various S proteins have been described for several viruses. In our study, we compared the efficiency of pseudotyped viruses with S proteins from different groups of CoVs. Furthermore, the cell tropisms of TGEV and PEDV were characterized by live and pseudotyped viruses.
-
293T cells were cultured and maintained in RPMI 1640 medium(Invitrogen, Carlsbad, CA, USA)supplemented with 10% fetal bovine serum(FBS)at 37 ℃ in an atmosphere of 5% CO2. PK-15, Huh-7, Vero-CCL-81, MDBK, CCL94, BSR, and MDCK cells were cultured and maintained in Dulbecco's Modified Eagle's modium(DMEM)(Invitrogen)supplemented with 10% fetal bovine serum(FBS)at 37 ℃ in an atmosphere of 5% CO2. The PEDV and TGEV strains(GenBank accession numbers: KT021232.1 for PEDV, HQ462571.1 for TGEV)were isolated from a suckling piglet. Rabbit anti-PEDV N protein and rabbit anti-TGEV N protein polyclonal antibodies are stored by the lab.
-
The S protein sequences of different groups of CoVs(GenBank accession numbers: ABD72982.1 for SARS-CoV-S, AAR92025.1 for MHV-S, NP_073551.1 for HCoV-229E-S, ABG89335.1 for TGEV-S, NP_598310.1 for PEDV-S, AAT84362.1 for HCoV-OC43-S, ABM66810.1 for BCoV-S, and NP_040831.1 for avian infectious bronchitis virus(IBV)S)were codon-optimized and synthesized(GenScript, Nanjing, China). The S fragments were cloned into the pcDNA3.1(+)vector with a C-terminal His-tag.
-
On the day before transfection, 293T cells were seeded in six-well plates. The next day, the cells were transfected with S plasmids plus the lentiviral vector HIV-Luc using the Lipofectamine 2000(Invitrogen)transfection reagent. After incubation for 5–6 h at 37 ℃, the supernatant of cultured cells was replaced by DMEM containing 2% FBS. After 48 h, the supernatants of transfected cells containing pseudotyped viruses were harvested, and the cell debris was removed by low-speed centrifugation. The pseudoviruses were quantified with an HIV-1 p24 EIA kit(Beckman-Coulter, Brea, CA, USA).
-
Lentiviruses pseudotyped with S proteins were solubilized by boiling in sodium dodecyl sulfate sample buffer. The S proteins were fractionated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The nitrocellulose membrane was incubated with a 1:4000 dilution of a mouse anti-His antibody(ProteinTech Group, Chicago, IL, USA). Goat anti-mouse IgG(Boster, Wuhan, China)was used as the secondary antibody.
-
A plasmid(Vector pcDNA 3.1(+))with each viral receptor gene(ACE2, CEACAM1, hAPN, and pAPN)was transfected into 293T cells with Lipofectamine 2000. After incubation for 5–6 h at 37 ℃, the supernatant was replaced by DMEM containing 2% FBS. After 48 h, the cells were infected by lentiviruses pseudotyped with the S proteins from CoVs of different groups.
-
For analysis of the lentiviruses pseudotyped with the S proteins from CoVs of different groups, different cell lines were seeded in 96-well plates. 293T cells expressing receptors were infected by the same lentiviruses and incubated at 37 ℃ for 2 h. DMEM(150 μL)containing 2% FBS was added subsequently to all cells and incubated for an additional 48–72 h. The pseudovirus entry efficiency was characterized based on luciferase activity.
-
PEDV and TGEV were used to infect various cell lines from different species, including PK-15(pig kidney), Huh-7(human liver), Vero-CCL-81(monkey kidney), MDBK(bovine kidney), CCL94(cat kidney), BSR(hamster kidney), and MDCK(canine kidney)cells at a multiplicity of infection of 0.01. Trypsin(10 μg/mL)was included in the cell culture medium to facilitate live virus infections. Cells infected with viruses were fixed with 4.0%(v/v)paraformaldehyde at 24 or 48 h post-inoculation. PEDV or TGEV was detected with fluorescein isothiocyanate-labeled rabbit anti-PEDV or anti-TGEV N protein antibodies, respectively, and observed under a fluorescence microscope.
-
All experiments were done at least twice(in most cases three or more times). Data were analyzed statistically using two-tailed student's t-tests for comparison of pcDNA3.1 and CoV S pseudovirus.
Cell lines, virus strains and antibodies
Plasmids
Production of lentiviruses pseudotyped with S proteins from CoVs of different groups
Western blot analysis of the pseudovirus construction
Expression of receptors
Infection of lentiviruses pseudotyped with S proteins
Cell lines were infected by PEDV and TGEV
Statistical analyses
-
CoV diversity is reflected in the variable S proteins(Heald-Sargent and Gallagher, 2012). Therefore, we compared the S protein amino acid sequences of CoVs from different groups, including SARS-CoV, MHV, HCoV-229E, TGEV, PEDV, HCoV-OC43, BCoV, and IBV(GenBank accession numbers ABD72982.1, AAR92025.1, NP_073551.1, ABG89335.1, NP_598310.1, AAT84362.1, ABM66810.1, and NP_040831.1, respectively)using ClustalW(Figure 1B). Because S1 is responsible for receptor binding, the amino acid sequences of the S1 domain were aligned(Figure 1C). The S proteins from CoVs of the same group shared sequence similarities of greater than 20%, with some similarities of up to 40%. The S1 sequence similarities of CoVs from different groups were less than 12%.
-
To determine the success of pseudovirus construction, the S proteins of pseudoviruses were confirmed by western blotting(Figure 2A). The pseudoviruses were quantified with an HIV-1 p24 EIA kit and used to infect cells expressing the corresponding receptor. Lentiviruses pseudotyped with SARS-CoV S protein could efficiently infect 293T cells expressing ACE2, and the pseudovirus level after entry reached 106 relative light units(RLU)(Figure 2B). The S proteins of HCoV-229E, TGEV, and PEDV exhibit high homology. However, their pseudovirus infection efficiency varies, and they are approximately 50-fold less efficient than the SARS-CoV-S pseudovirus. The TGEV-S and PEDV-S pseudo-viruses showed similar infection efficiencies. Although HCoV-OC43, BCoV, and IBV utilize a sugar as their receptors, their pseudovirus infection efficiency was not clear.
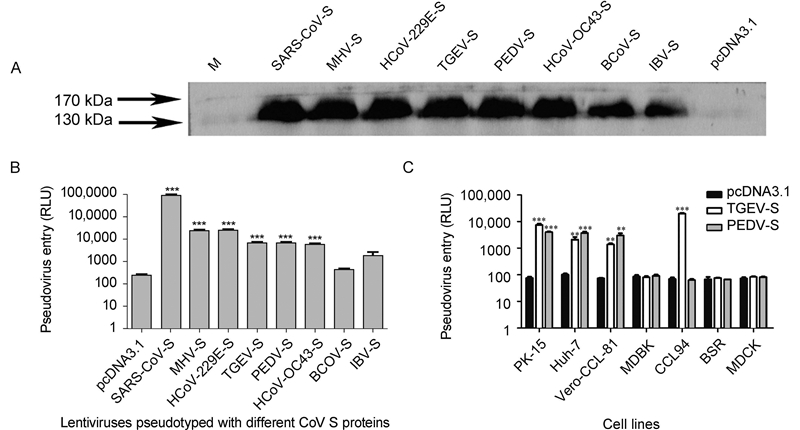
Figure 2. Coronavirus-spike-mediated entry of pseudoviruses into host cells. (A) Western blotting of the S protein of pseudoviruses. (B) Lentiviruses pseudotyped with different coronavirus S proteins were generated and used to infect different cells. SARS-CoV-S pseudoviruses infected 293T cells expressing endogenous ACE2. TGEV-S and PEDV-S pseudoviruses infected 293T cells expressing endogenous pAPN. MHV-S and HCoV-229E-S pseudoviruses entered cells expressing CEACAM1 and hAPN, respectively. BCoV-S, HCoV-OC43-S, and IBV-S infected 293T cells directly. (C) TGEV-S and PEDV-S pseudoviruses infected PK-15, Huh-7, Vero-CCL-81, MDBK, CCL94, BSR, and MDCK cells. The pseudovirus entry efficiency is reported as the luciferase activity. Statistical analyses were performed to compare the treatment groups: ***P < 0.001; **0.001 < P < 0.01.
TGEV-S and PEDV-S pseudoviruses were used to infect PK-15, Huh-7, Vero-CCL-81, MDBK, CCL94, BSR, and MDCK cells(Figure 2C). The TGEV-S and PEDV-S pseudoviruses could enter PK-15, Huh-7, and Vero-CCL-81 cells with a similar efficiency. However, there was an intriguing finding that CCL94 cells could be infected efficiently by TGEV-S but not PEDV-S pseudoviruses.
-
To further study the cellular entry of CoVs, we used live PEDV and TGEV to infect different cell lines. PEDV efficiently infected Vero-CCL-81(monkey kidney), Huh-7(human liver), and PK-15(pig kidney)cells(Figure 3). Infection was not evident in MDBK(bovine kidney), CCL94(cat kidney), BSR(hamster kidney), or MDCK(canine kidney)cells. This is consistent with the data in Figure 2C. TGEV efficiently infected cells from pigs(PK-15), humans(Huh-7), monkeys(Vero-CCL-81), and felines(CCL94)(Figure 4). Overall, our study demonstrates that PEDV and TGEV can infect cells from different species.
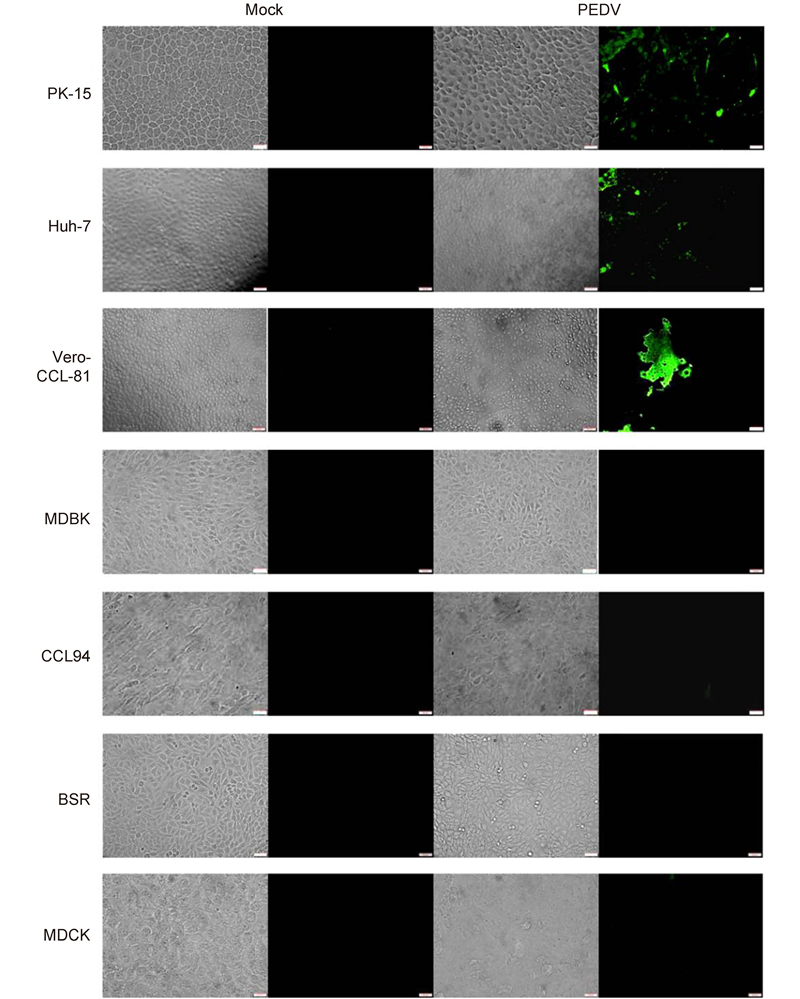
Figure 3. PEDV infections in cultured cells. The PEDV strain, YN144, was used to infect different cell lines at a multiplicity of infection of 0.01. Trypsin (10 μg/mL) was included in the medium to facilitate live PEDV infection. Cells infected with PEDV were fixed with 4.0% (v/v) paraformaldehyde at 24 h post-inoculation. PEDV was detected with a fluorescein isothiocyanate-labeled rabbit anti-PEDV N protein antibody and observed under a fluorescence microscope. Scale bars are 50 μm.
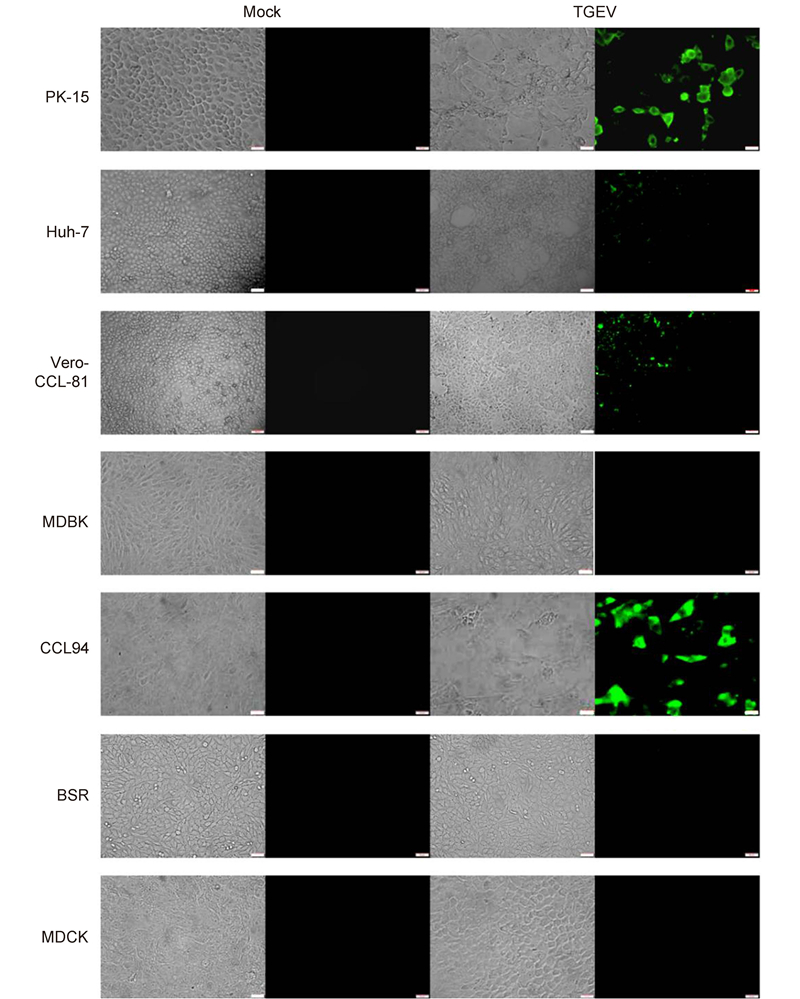
Figure 4. TGEV infections in cultured cells. The TGEV-WH-1 strain was used to infect different cell lines at a multiplicity of infection of 0.01. Trypsin (10 μg/mL) was included in the medium to facilitate live TGEV infections. Cells infected with TGEV were fixed with 4.0% (v/v) paraformaldehyde at 48 h post-inoculation. TGEV was detected with a fluorescein isothiocyanate-labeled rabbit anti-TGEV N protein antibody and observed under a fluorescence microscope. Scale bars are 50 μm.
Sequence alignment
Comparison of lentiviruses pseudotyped with S proteins of CoVs
Live PEDV and TGEV have different cell tropisms
-
CoVs recognize a variety of cell-surface molecules as their host receptors, including proteins, sugars, and heparan sulfate(Li, 2013). The cell entry mechanisms of these viruses are mediated by the viral S proteins that bind cellular receptors and mediate the fusion of viral and cellular membranes(Heald-Sargent and Gallagher, 2012). We demonstrated that S proteins from CoVs of different groups played an important role in mediating viral infection at different efficiencies. SARS-CoV-S pseudoviruses efficiently entered cells expressing ACE2. This result is consistent with previous reports demonstrating that ACE2 is a receptor for SARS-CoV(Schwegmann-Wessels et al., 2009; Wu et al., 2011). HCoV-229E-S and MHV-S pseudoviruses showed strong infectivity, in part because they use proteins as a receptor(Li et al., 2005; Belouzard et al., 2012). Moreover, PEDV-S and TGEV-S pseudoviruses had similar infection efficiencies in PK-15, Huh-7, or Vero-CCL-81 cells, and TGEV-S pseudov-irus-infected CCL94 cells.
The sugar moiety 5-N-acetyl-9-O-acetylneuraminic acid(Neu5, 9Ac2), found on cell-surface glycoproteins or glycolipids, is recognized by BCoV and HCoV-OC43. In addition, two other types of sugars, 5-N-glycolylneuraminic acid(Neu5Gc) and 5-N-acetylneuraminic acid(Neu5Ac), can serve as receptors or co-receptors for some alpha-and gamma-CoVs(Cavanagh and Davis, 1986; Krempl et al., 1997; Peng et al., 2012; Shahwan et al., 2013). The S-to-sugar binding affinity is lower than the S-protein interaction, which partially explains why the BCoV and IBV pseudoviruses have low entry efficiency.
Although PEDV and TGEV use the same receptor(pAPN) and the S proteins present high homology, live PEDV and TGEV prefer different cell lines; PEDV prefers Vero-CCL-81, Huh-7, and PK-15 cells. This is consistent with a previous report(Liu et al., 2015). However, TGEV prefers Vero-CCL-81, Huh-7, PK-15, and CCL94 cells. This indicates that PEDV and TGEV may have different species preferences. These results are consistent with the observation that their receptor-binding domains are located in non-homologous regions(Belouzard et al., 2012). However, the mechanism underlying different cell tropisms requires further investigation.
The S protein plays a crucial role in entry into host cells by mediating receptor binding and membrane fusion. CoVs use a variety of receptors and triggers to activate fusion(Belouzard et al., 2012). The first and most important step of virus infection is the interaction between the virus and its cellular receptor. Infection of lentiviruses pseudotyped shows that S proteins from different CoVs have varied abilities to mediate pseudotyped virus infection in different cell types from different tissues. These findings further our understanding of the mechanisms underlying viral invasion and contribute to the development of drugs against CoVs.
-
This work was supported by the National Natural Science Foundation of China(Grant No. 31372440).
-
This article does not contain any studies with human or animal subjects performed by any of the authors. There is no competing interest in this research.
-
G.P. and J.W. designed the experiments and wrote the paper. J.W. performed the experiments. F.D. and Q.H. contributed to critical discussions of the data. All authors approved the final manuscript.














 DownLoad:
DownLoad: