-
Human cytomegalovirus (HCMV) is a ubiquitous pathogen with high infection rate throughout the world. Associated with socio-economic status, the HCMV sero prevalence among adult varies from 36%–77% in developed areas such as North America and Europe to around 95% in developing country including China (Adland et al., 2015). Besides contributing to diseases in immunocompromised hosts, HCMV is also a leading cause of congenital infections resulting birth defects, primarily maldevelopment of the central nervous system (CNS) (Luo et al., 2010). Each year approximately 1% to 2% of all newborns are congenitally infected with HCMV, and 5% to 10% of these infants manifest signs of serious neurological defects (Boppana et al., 1992; Britt and Mach, 1996; Cinque et al., 1997; Fowler et al., 1997). An additional 10% of congenitally infected infants are asymptomatic at birth but subsequently develop brain disorders, the most common of which is sensorineural hearing loss (Pass et al., 1980; Conboy et al., 1986; Qiao et al., 2016).
Although HCMV may infect multiple tissues in vivo (Sinzger and Jahn, 1996), its principal site of manifestations is the subventricular zone (SVZ) in the fetal brain (Luo et al., 2008). The major cell type and primary target of HCMV infection in SVZ is the neural progen itor/stem cells (NPCs), which are also the bases for neuronal development (Luo et al., 2010). Therefore, NPCs is used as an ideal model to investigate the pathogenesis of HCMV induced neural developmental disorder and to reveal the underlying mechanism (Luo et al., 2008; Pan et al., 2013).
Our previous works have shown that NPCs are fully permissive for HCMV infection (Luo et al., 2008). HCMV infection suppresses the self-renew of NPCs and causes their abnormal differentiation, implying a possible mechanism for the CNS manifestations of HCMV pathogenesis (Luo et al., 2010). Moreover, NPCs at later passages are more permissive and sensitive to HCMV infection, which may cause more neural cell loss and give rise to severer neurological disabilities with advanced brain development (Pan et al., 2013). In spite of the clear evidences suggesting congenital HCMV infection causes neural maldevelopment through influencing NPCs, the detailed mechanism remains unclear.
Normal neural development requires proper NPCs’ self-renewing and differentiation, and Notch signaling pathway plays an essential role in regulating NPCs cell fate (Gaiano and Fishell, 2002; Selkoe and Kopan, 2003; Shimojo et al., 2008). Multiple factors are involved in Notch signaling, including ligands (such as Delta and Jag), receptors (Notch), mediators (RBP-j), and effectors (Hes) (Fortini, 2009). Binding of Notch signaling ligands and receptor causes the splicing of Notch receptor, releases the Notch intracellular domain (NICD), which forms a complex with RBP-J and activates the expression of the downstream effector genes, one important member of which is Hes1 (Honjo, 1996; Selkoe and Kopan, 2003).
Hes1 expression in NPCs is fine-tuned by complicated regulation mechanism. Besides the transactivation through upstream signaling of Notch pathway, the transcription of Hes1 is also repressed by the direct suppressive binding of Hes1 protein to the N box sequences in its own promoter (Takebayashi et al., 1994). The Hes1 protein undergoes rapid degradation through ubiquitination and proteasome pathway, resulting the depletion of Hes1 protein. Then, the disappearance of Hes1 protein releases its suppression on Hes1 promoter, and initiates the next round of Hes1 expression (Hirata et al., 2002; Kageyama et al., 2009). The feeding back inhibition and fast protein degradation cause the unique oscillatory expression pattern of Hes1 in NPCs, whose period is about 2 hour in mouse NPCs (Masamizu et al., 2006). However, the periods of Hes1 oscillation is not stable but varied from cycle to cycle and from cell to cell (Shimojo et al., 2008). Cell cycle progression is irrelevant to Hes1 rhythm, since inhibiting DNA replication by Ara-C doesn’t influence the oscillation in cells (Hirata et al., 2002).
As the essential effectors of Notch signaling, the proper oscillatory expression of Hes1 plays an essential role in maintaining the stemness of NPCs. The sustained expression of Hes1 after losing its rhythm drives NPCs enters dormant, a state neither proliferating nor differentiating (Kageyama et al., 2009). In Hes1 knock-out mice, many radial glial cells are not well maintained and undergo prematured differentiation into neurons, which in turn results in accelerated neuralgenesis (Ishibashi et al., 1995; Cau et al., 2000). Neurospheres of NPCs with Hes1 deficiency also fail to properly expand (Ohtsuka et al., 1999). Moreover, inactivation of Hes1 upregulates the expression of proneural genes and accelerates neuronal differentiation (Tomita et al., 1996; Hatakeyama et al., 2004). Therefore, dysregulation of Hes1 expression leads abnormal NPCs differentiation and may further cause neural developmental disorders. However, all of the Hes1 knowledge is obtained from mouse model, and few data is available of Hes1 in human neural system. The relationship between HCMV and Hes1 rhythm remains to be revealed.
Our previous studies have demonstrated that HCMV infection dysregulate Notch signaling pathway by downregulating the main ligand (Jag1) and effective receptor NICD1 in human NPCs (Li et al., 2015), which substantially contributes in HCMV induced NPCs premature and abnormal differentiation in some extent. Since Hes1 is an important downstream effector in Notch signaling, it is of great interest to investigate whether HCMV infection also influences Hes1 in human NPCs. In the present study, Hes1 expression rhythm was proved for the first time to exist in naïve human NPCs, with a period of about 1.5 hour. HCMV infection disrupted this natural rhythm and decreased Hes1 level. Knocking down Hes1 with specific siRNA suppressed the proliferation of monolayer NPCs and neurosphere formation. In addition, the four important NPCs hallmarks, Sox2, DCX, GFAP, and Nestin were also dysregulated, indicating the abnormal cell fate of NPCs. Altogether, these data suggest that HCMV infection affects NPCs self-renew and cell fate potentially through breaking Hes1 rhythm and decreasing its expression.
-
NPCs and human embryonic lung fibroblast cells (HELs) were isolated and cultured as previously described (Pan et al., 2013). Briefly, NPCs were proliferated in fibronectin (14.28 μg/mL, Millipore) coated tissue culture dishes, then digested with accutase (Millipore) and transferred to poly-D-lysine (50 μg/mL, Millipore) coated dishes for further experiments. NPCs growth medium (GM) was Dulbecco’s modified Eagle’s medium-F12 (DMEM-F12) containing amphotericin B (1.5 μg/mL), L-glutamax (2 mmol/L), penicillin-streptomycin (100 U/mL), Gentamicin (50 μg/mL), human basic fibroblast growth factor (bFGF, 20 ng/mL), human epithelial growth factor (EGF, 20 ng/mL) (Gibco) and 10% BIT9500 serum substitute (Stem Cell Technologies). The medium for NPCs culture was half changed with fresh GM every two days, and the old medium was collected as condition medium (CM).
For the culturing of HELs and 293T cells (ATCC #CRL-321), Minimum essential medium (MEM) and Dulbecco modified Eagle medium (DMEM) were utilized, respectively, with the supplementation of 10% fetal bovine serum (Gibco/Life Technology), glutamine (2mmol/L, Gibco/Life Technology), and penicillin-streptomycin (100 U/mL and 100μg/mL). Cells were cultured in appropriate medium at 37 °C in a humidified atmosphere containing 5% CO2.
-
HCMV Towne strain (ATCC-VR977) were propagated in HEL as described previously (Fu et al., 2015). Briefly, HEL (6 × 106) were infected with the virus at a multi plicity of infection (MOI) of 0.02, and the supernatants were harvested at 7 and 10 days post infection (dpi). Viruses were concentrated by ultracentrifugation at 4 °C (25, 000 × g for 4 h), resuspended in GM containing 1% dimethyl sulfoxide (DMSO), titrated by plaque assay as described previously (Casavant et al., 2006), and stored at –80 °C as aliquots.
For HCMV infection, NPCs were cultured in GM without BIT9500 for 24 h to synchronize the cell status. Then, the synchronized cells were infected by virus at an MOI of 3. 3 h later, the inoculum soup was replaced with culture medium consisting of fresh GM and CM at equal volume. Cells were harvested for further assays at the indicated time points.
-
Total RNA was isolated from the harvested cells using Trizol Reagent (Takara), quantitated by NanoDrop 2000 Spectrophotometer (Thermo Co., USA), and reverse-transcribed using PrimeScript RT Master Mix (TaKaRa). Quantitative real-time PCR reactions were performed using iTaq Universal SYBR Green Supermix on a CFX Connect Real-Time PCR Detection System (Bio-Rad). Each sample was run in duplicate with GAPDH as the normalizing reference. The primers for qRT-PCR is as following: Hes1, F-ACCCCAGCCAGTGTCAAC, R-GAATGTCCGCCTTCTCCA; GAPDH, F-GAAGGTGAAGGTCGGAGTC, R-GAAGGTGAAGGTC GGAGTC.
Primers Sequence scramble Forward Primer (5′-3′) UUCUCCGAACGUGUCACGUTT Reverse Primer (5′-3′) ACGUGACACGUUCGGAGAATT Hes1-1 Forward Primer (5′-3′) CGGCCAAUUUGCCUUUCUCTT Reverse Primer (5′-3′) GAGAAAGGCAAAUUGGCCGUC Hes1-2 Forward Primer (5′-3′) CGACACCGGAUAAACCAAATT Reverse Primer (5′-3′) UUUGGUUUAUCCGGUGUCGUG Hes1-3 Forward Primer (5′-3′) CACUGAUUUUGGAUGCUCUTT Reverse Primer (5′-3′) AGAGCAUCCAAAAUCAGUGTT Hes1-4 Forward Primer (5′-3′) GCCUAUUAUGGAGAAAAGATT Reverse Primer (5′-3′) UCUUUUCUCCAUAAUAGGCTT Table 1. Primers for siRNA
-
Cells were harvested with cell scraper, pelleted by centrifugation, resuspended in ice-cold PBS, snap frozen in liquid nitrogen, and stored at –80 °C. The cells then were lysed in RIPE buffer (50 mmol/L Tris-base, 1.0 mmol/L EDTA, 150 mmol/L NaCl, 0.1% SDS, 1% tritonX-100, 1% sodium deoxycholate and cOmplete Protease In hibitor [Roche]). The protein concentration was measured by the BCA kit (Beyotime) according to the manufacturer’s protocol. Samples with equal amount of protein were separated by SDS-PAGE gel and trans ferred to PVDF membranes (0.25 μm, Millepore). After incubation with the indicated primary and corresponding secondary antibodies, signals were detected using a Chemiluminescence machine, and analyzed by den- sitometry program (Image J). At least three sets of in dependent experiments were performed and repre- sentative results were shown. The primary antibodies used included mouse monoclonal antibodies against -Hes1 (IgG1, Abcom), -GFAP (IgG2b, Fitzgerald In dustries International, Inc.), -Nestin (IgG1; Chemicon), -SOX2 (cell signal, IgG1), -DCX (IgG; Santa Cruz Biotechnology) and β-actin (Santa Cruz Biotechnology). Secondary antibodies used were horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Amersham Bioscience).
-
To determine the half-life of Hes1 protein (t1/2), NPCs were treated with 0.1mg/mL cycloheximide (CHX, Sigma-Aldrich) (Ajiro and Zheng, 2015) and collected at the indicated time points, Hes1 protein levels were enriched by Immunoprecipitation (IP), and then examined by IB. Hes1 degradation curve was generated by the log2 of the relative Hes1 protein level (Hes1/β-actin) normalized to that of 0 min (y) versus sampling time (x). The half-life of endogenous Hes1 protein were measured and calculated as previously described (Hirata et al., 2002).
-
siRNAs were synthesized at GenePharma company, and the sequences were listed in Table 1. 293T cells transfected with Hes1 expressing construct (pCDH-Hes1) applied to assess the knocking down efficiency of the siRNAs. Briefly, 4 × 106 293T cells were seeded in 6-well plate one day prior to transfection. 100 pmoL siRNA and pCDH-Hes1 or 5 μL Lipofectamine (Invitrogen) were diluted to a total volume of 50 μL, respectively. After 5 minites incubation, the diluted DNA and Lipofectamine were mixed, and incubated for 20 minutes at room temperature. The mixture was then added to 293T cells and the medium was changed 4 h later. Cells were harvested 48 h later to examine the Hes1 protein amount.
siRNA were introduced to NPCs by Nucleofection, which was performed using Amaxa Mouse NSC Nucleofector Kit (Lonza) following the manufacture's instruction. Briefly, 110 μL reaction medium was prepared by mixing Nucleofector solution (82 μL), supplement (18 μL) and siRNA (scr and si-3, 100 pmol). NPCs were resuspended in the reaction medium, transferred into a certified cuvette, applied to Nucleofector Program A-033, NPCs were then cultivated in uncoated dishes as neurospheres or in poly-D-lysine-coated dishes as monolayers (Li et al., 2015).
-
NPCs nucleofected with siRNA were cultured on uncoated or poly-D-lysine-coated culture dishes for 48 h. Then the images were obtained using a Nikon Eclipse TS100 inverted microscope equipped with a Nikon CoolPix P6000 camera. Representative images were collected from three independent experiments. The monolayer NPCs on coated surface were digested with accutase for 1 to 2 min, pelleted by centrifugation, resuspended and counted using a cell-count board. The neurospheres were categorized by diameter into three size groups (small, < 50 μm; medium, 50–100 μm; and large, > 100 μm).
-
All the results are representative of at least three in-dependent experiments. Differences were considered statistically significant at P ≤ 0.05 based on a Student’s t-tests analysis.
-
Hes1 expression rhythm has been observed in various cultured cells, such as fibroblast, myoblasts, neuro-blastoma cells, teratocarcinoma cells as well as mouse NPCs (Hirata et al., 2002; Shimojo et al., 2008). Os-cillatory Hes1 expression on most cultured cell lines was triggered by serum starvation followed by serum stimulation in the indicated cells (Hirata et al., 2002). Whereas the in vivo Hes1 expression rhythm was measured in isolated mouse NPCs after transfecting the mouse brain using the luciferase reporter under the control of Hes1 promoter (Shimojo et al., 2008). However, to our knowledge, Hes1 rhythm has not been demonstrated in human NPCs so far.
To examine whether Hes1 oscillatory expression also exists in naïve human NPCs, cells were starved in BIT9500-free growth medium (GM) for 24 h prior to be culture in GM containing BIT9500, and then collected every 0.5 h. To measure the Hes1 level, cells were harvested every 0.5 h since the administration of BIT9500 up to 5.5 h, and subjected to quantitative reverse transcriptase PCR (qRT-PCR). The relative Hes1 mRNA levels at different time points were presented as the fold change to 0 h after normalizing to the internal control GAPDH. Considering the slight delay of protein synthesis to mRNA transcription, samples for immunoblotting were collected starting at 0.25 h (Hirata et al., 2002). The relative Hes1 protein level was quantitated according to the immunoblotting (IB) results, and presented as the fold change to 0.25 h with β-actin normalization. The oscillatory rhythm of Hes1 expression, both at mRNA and protein levels, was clearly observed (Figure 1A, 1B). The peak levels of Hes1 mRNA appeared at 1.5, 3 and 4.5 h, indicating the transcription period of ~1.5 h (Figure 1A). The Hes1 protein levels peaked at 0.75 h, 1.75~2.75, 4.25, 5.75 and 7.25 h, confirming a ~1.5 h periodic cycle (Figure 1B). Subsequently, the half-life (t1/2) of Hes1 protein in NPCs was investigated by monitoring the steady-state levels following treatment with the protein synthesis inhibitor cycloheximide (CHX) for 30 min, 60 min and 90 min (Hirata et al., 2002). Since the endogenous Hes1 level is very low in NPCs, the protein was enriched by immunoprecipitation (IP) prior to IB. The t1/2 of the Hes1 protein in NPCs was 17.6 ± 0.2 min (Figure 1C).
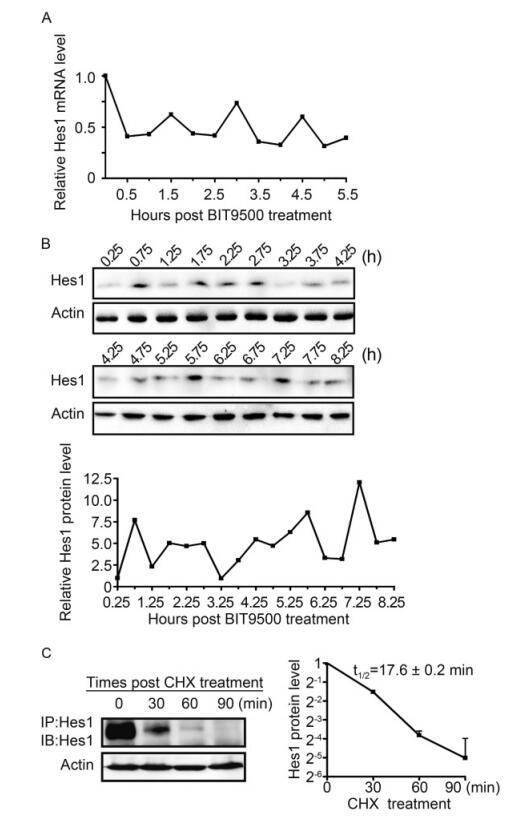
Figure 1. Hes1 exhibits oscillatory expression in naive human NPCs. (A) Hes1 oscillation at the mRNA level. NPCs were cultured in BIT9500-free GM for 24 h before being switched to regular GM (containing BIT9500). At the indicated times post BIT9500 stimulation (0 h), the mRNA levels of Hes1 relative to GAPDH were quantified by qRT-PCR and normalized to the mRNA levels at 0 h (set to 1.0). Shown is the representative result from 3 independent experiments. (B) Hes1 oscillation at the protein level. NPCs were cultured as described in (A). At the indicated times after BIT9500 treatment (0 h), the protein levels of Hes1 relative to p-actin were assessed by im- munoblotting (upper panel) and quantified by densitometry, where relative Hes1/Actin signal ratios were normalized to that at 0.25 h (set to 1.0) (lower panel). Shown is the representative result from 3 independent experiments. (C) Halflife of Hes1 protein in naive NPCs. NPCs were cultured in BIT9500-containing GM and treated with CHX for the indicated times. Following enrichment by Hes1-directed IP from 3 × 107 NPCs, the protein levels of Hes1 relative to P-actin were assessed by IB (left panel) and quantified by densitometry, where signal ratios of Hes1/Actin were normalized to the signals at the start (0 min) of CHX treatment (set to 1.0). Results are presented as means with standard deviations from three independent experiments. The average half-life (t1/2) of Hes1 protein is calculated as the x value when y = 0.5 using the formula: Log2(y) = -0.0442x+0.0898, R2 = 0.9945, where x is the CHX treatment time and y is the relative Hes1 protein level (right panel).
These data demonstrate for the first time the existence of Hes1 expression rhythm, with about 1.5 h periodic cycle, in human NPCs, at both the mRNA and protein level.
-
Hes1 rhythm has been observed in a wide range of cell types, but whether virus infection has any influence on it remains unclear. Our previous published data have shown that human NPCs are fully permissive for HCMV infection (Luo et al., 2008; Luo et al., 2010), so NPCs/HCMV model was applied to study the relationship between HCMV and Hes1 rhythm.
Starving synchronized NPCs were infected with HCMV Towne strain at an MOI of 3, with the mock infection as control. Hes1 mRNA levels were examined at 30 minutes interval during the immediate early (IE), early(E), early-late (EL) and late stages (L) of HCMV infection. In mock-infected NPCs, the rhythm of Hes1 mRNA with 1.5 h period was clearly observed even up to 2 days, and the average Hes1 mRNA level remained similar as the baseline (t = 0) (Figure 2B–2D) in spite of a burst immediate after the BIT9500 stimulation (Figure 2A). On the contrary, Hes1 mRNA rhythm in HCMV infected NPCs matched that in mock control in the first 3 hours post infection, and asynchonization appeared since 3.5 hpi indicating that the Hes1 mRNA rhythm started to be altered (Figure 2A). Then the Hes1 mRNA rhythm was gradually dampened during early and early-late stage (Figure 2B , 2C), and finally disappeared in late stage (Figure 2D). Notably, not only the rhythm was disrupted by HCMV infection, but also Hes1 mRNA level in NPCs was influenced upon HCMV infection. Compared to mock control, Hes1 mRNA level in HCMV infected NPCs kept decreasing since 3.5 hpi (Figure 2A–2D), until finally reached the sustained but low level, which dropped to about 25% of that at t = 0 (Figure 2D).
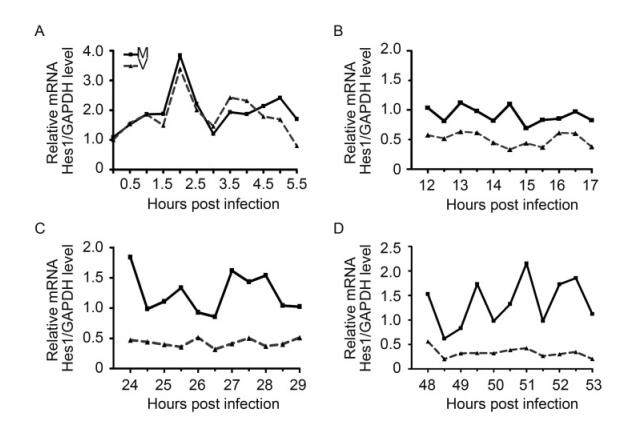
Figure 2. HCMV infection disrupts Hes1 mRNA rtiythm. Synchronized NPCs were infected with HCMV at an MOI of 3 (dotted line), and mock infection (solid line) was applied as control. Cells were harvested at 0.5 h interval at the immediate early (A), early (B), early-late (C) and late stages (D). Relative Hes1 mRNA level against GAPDH was quantified by qRT-PCR and normalized to that of 0 hpi. Shown is the representative result from 3 independent experiments.
Because of the relative short half-life of Hes1 protein in NPCs, which is about 17.6 min (Figure 1C), the alteration on Hes1 mRNA level should also lead changes on Hes1 protein level. This hypothesis prompted us to examine the Hes1 protein level in NPCs with or without HCMV infection. Starving synchronized NPCs were infected with HCMV Towne strain at an MOI of 3, with mock infection as control. Hes1 protein levels were firstly examined during the IE stage with 30 min interval at 0.25–3.75 hpi and 1 h interval at 4–11 hpi, with β-actin as the internal loading control. The IB for infected and mock samples at the same time points was performed on the same blot, and the images were split and displayed in parallel for direct comparison (Figure 3A, upper panel). According to the densitometric quantification result, Hes1 protein level maintained the oscillated pattern with similar period at the IE stage of HCMV infection. The frequency of Hes1 oscillation after HCMV infection was similar to that of mock infected cells, the average Hes1 protein amount, as well as the peak level and valley level, remains similar during the IE stage but the phase is different. These data demonstrate that Hes1 protein rhythm displayed certain level of asynchronous compared to mock (Figure 3A, lower pannel). The rhythm asynchronization and similar level of Hes1 protein at IE stage are consistent with the phenomena observed at mRNA level (Figure 2A).
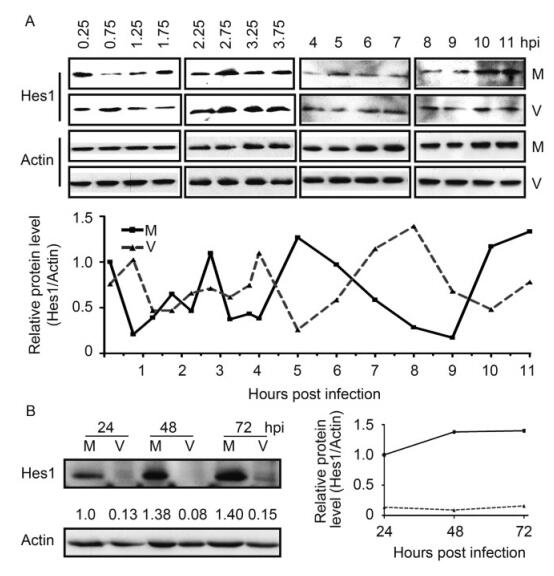
Figure 3. HCMV infection disrupts Hes1 protein rhythm. (A) Hes1 protein oscillation at IE stage of HCMV infection. Synchronized NPCs were infected with HCMV at an MOI of 3 (V), and mock infection (M) was applied as control. Cells were harvested at the indicated times and subjected to IB to examine the protein level of Hes1. p-actin serves as an internal loading control (upper panels). The representative IB images from three independent experiments were shown, and the protein levels of Hes1 relative to β-actin were also quantified by densitometry (lower panel). (B) Hes1 protein level is downregulated by HCMV versus mock infection. Following mock infection (M) or infection with HCMV (V) at an MOI of 3, NPCs were collected at the indicated times. β-actin served as loading control, the protein levels of Hes1 relative to β-actin were also quan-tified by densitometry.
The low baseline level of Hes1 protein required large amount of NPCs to detect Hes1 protein by IB. And the difficulties of obtaining and expanding NPCs challenged the attempts to examine the Hes1 protein level with less interval. Therefore, HCMV infected NPCs were harvested at 24, 48 and 72 hpi to examine the Hes1 protein level. The sampling frequency is not enough to determine the rhythm, but the Hes1 protein level was dramatically decreased compared to the corresponding mock control (Figure 3B), which matched the alteration of Hes1 mRNA at the same stage in HCMV infected NPCs (Figure 2C–2D).
During the IE stage post infection, HCMV altered the oscillation phase of Hes1 protein, but didn’t display influences on the average Hes1 protein amount, as well as the rhythm period and peak/valley levels. At 24 hpi and the later time points, Hes1 protein level was dramatic decreased in the HCMV infected NPCs compared to mock control. The above presented results clearly demonstrated that HCMV infection disrupts the Hes1 rhythm and down-regulates Hes1 level at both mRNA and protein levels, and this effect was achieved gradually instead of abruptly.
-
To clarify the physiological significance of Hes1 rhythm disruption and down-regulation, we knocked down Hes1 using siRNA. Because of the low baseline expression level of Hes1 in NPCs, the knock-down efficiency of 4 different Hes1 specific siRNAs were first assessed using exogenous Hes1. 293T cells were cotransfected with Hes1 expressing construct and specific siRNA (si-1 to si-4) or a scrambled siRNA control (scr) respectively. Hes1 protein level was analyzed at 48 h post transfection with β-actin as loading control. The result showed that si-2, si-3 and si-4 all efficiently decreased Hes1 protein level, while si-3 displayed the strongest effect (Figure 4A). Si-3 also efficiently depleted Hes1 protein level in NPCs (Figure 4F). Therefore, si-3 was applied in future studies to knock down Hes1.
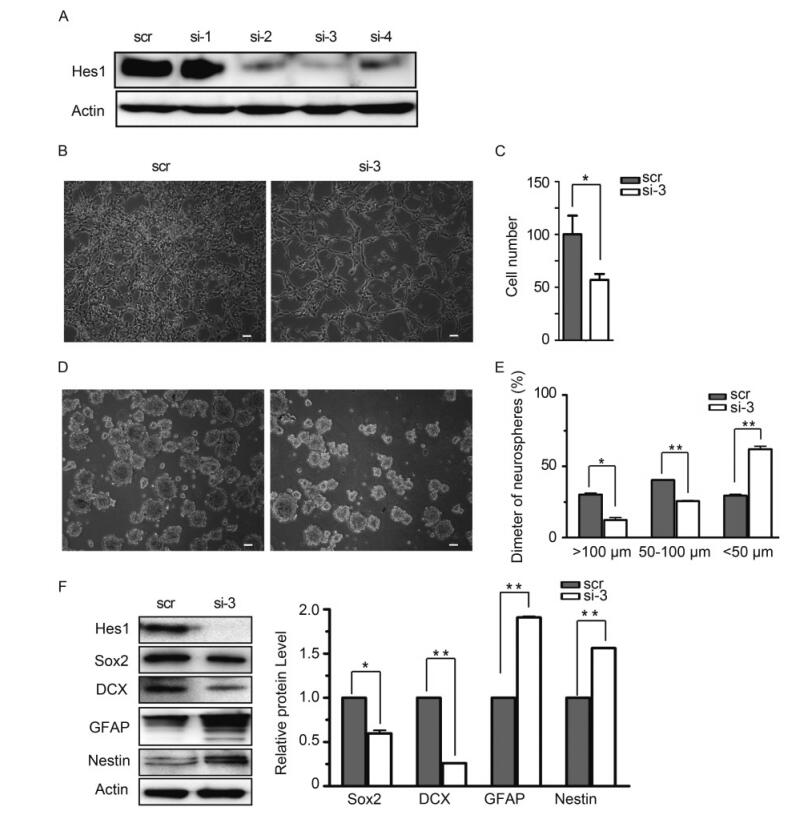
Figure 4. Knocking-down Hes1 affects proliferation of monolayer NPCs and halts the growth of human neurospheres. (A) Knock-down efficiency of candidate Hes1 siRNA in 293T cells. 293T cells were transfected with siRNA-Hes1-1 (si-1), siRNA-Hes1-2 (si-2), siRNA-Hes1-3 (si-3), siRNA-Hes1-4 (si-4) or siRNA-scramble (scr) together with Hes1 expression construct. Hes1 protein levels were examined by IB 48 h later, with β-actin as control. (B-C) Proliferation comparison. NPCs nucleofected with si-3 or scr were cultured on coated surface, images were obtained after 48 h culture, and cells were digested and counted afterwards. Numbers of NPCs nucleofected with si-3 were normalized to the numbers of NPCs nucleofected with scr (set to 100). Shown are the representative images from three independent experiments (B), and the average results are presented as the mean ± SD (C). Scale bar, 50 μm. (D-E) Comparison neurospheres formation. NPCs nucleofected with si-3 or scr were cultured on uncoated surface to form neurospheres, and images were obtained after 48 h culture. The neurospheres were categorized into three size groups (small, < 50 μm; medium, 50–100 μm; and large, > 100 μm). Shown is the representative image from three independent experiments (D). For each experiments, three random images were obtained and analyzed, and the average result from all three experiments are presented as the mean ± SD (E). Scale bar, 50 μm. (F) Comparison of NPCs markers’ expression. NPCs nucleofected with si-3 or scr were cultured on uncoated surface for 48 h, and harvested to examine the indicated proteins by IB. The relative protein level were normalized according to β-actin after densitometry quantification, Shown is the representative image and the quantification data from three independent experiments. *, P < 0.05; **, P < 0.01.
To compare the effect of Hes1 decrease in NPCs, the cell proliferation ability, neuralsphere formation capacity, as well as the typical marker expression levels were examined in NPCs nucleofected with Hes1 specific (si-3) or scramble siRNA (scr). When equal amount of the nucleofected NPCs (2 × 106) were cultured as monolayers in poly-D-lysine-coated dishes (Figure 4B), the scramble control group (scr) proliferated to 4.1 × 106 ± 3.4 × 105 cells 2 days later, but Hes1 knocking down group only reached to 2.4 × 106 ± 5 × 104 cells (Figure 4C). The neuralsphere formation capacity of NPCs were examined by culturing equal amount of cells (2 × 106) on uncoated surface. 2 days later, the formed neurospheres were categorized into three size groups by diameters (small, < 50 μm; medium, 50–100 μm; and large, > 100 μm) (Figure 4D). Quantitative analysis demonstrated that the neuralspheres formed by control NPCs (scr) distributed evenly in the three size groups, 30.2% ± 1.3% large, 40.3% ± 0.1% medium, and 29.4% ± 1.4% small. In contrast, much less large (12.3% ± 2.4%) and medium (25.6% ± 0.6%) sized neurospheres were formed by NPCs after Hes1 knocking down (si-3), and majority of the neuralspheres (62% ± 3.1%) had a diameter smaller than 50 μm (Figure 4E). Hence, depleting Hes1supressed the proliferation and neuralsphere formation of NPCs.
To determine whether Hes1 depletion also altered the expression of typical NPC markers, Sox2, DCX, GFAP and Nestin protein level were assayed by IB with β-acting as the loading control. Compared to control (scr), Hes1 depleted NPCs (si-3) expressed significantly decreased amount of Sox2 and DCX, but GFAP and Nestin were upregulated (Figure 4F). This NPCs marker alteration pattern matched neither glia nor neuron differentiation profile of normal NPCs, indicating that depletion of Hes1 drove NPCs abnormal differentiation toward a non-glial and non-neuronal lineage.
Taken together, these results demonstrate that knocking down Hes1 by siRNA suppressed NPCs neuralphere formation capacity, but also prompted NPCs abnormal differenciation.
-
It has long been established that congenital infection causes neurological disorders such as microencephaly, hydrocephaly, hearing loss and mental retardation (Goderis et al., 2014). More recently, HCMV was also implicated to be related with autism (Pass et al., 1980; Yamashita et al., 2003).These disorders are characterized by either, or both, structural and functional abnormalities in the brain. There are at least four basic processes that determine brain structure and function: (1) proliferation of NPCs controlling neural cell numbers; (2) differ-entiation of NPCs controlling the numbers and function of neural cells, including mature neurons and glia; (3) migration of differentiated neural cells controlling brain structure; and (4) synapse and network formation among mature neurons and glia controlling the structural and functional brain connectome (Guerrini and Parrini, 2010; Hofman, 2014; Biran et al., 2015). However, which of these processes are affected by HCMV and how the virus may target them to cause neuropathy are incompletely understood. By using human NPCs as a model, we previously demonstrated that HCMV perturbs cell pro-liferation and differentiation, and we started focusing on the mechanisms underlying these perturbations (Luo et al., 2008; Koh et al., 2009; Luo et al., 2010; Pan et al., 2013).
Although having been confirmed in mouse NPCs, the existence of Hes1 rhythm remains obscure in human NPCs. The oscillatory period of Hes1 expression is 2–3 hours in mouse NPCs and embryonic brain tissues, which is different from those of other cultured cell lines after serum treatment (Masamizu et al., 2006; Shimojo et al., 2008). Limited by the ethics, low baseline expression level and short half-life, measuring Hes1 rhythm in human NPCs encounters great difficulties. In the present study, we reported for the first time that Hes1 expression also oscillates in naïve human NPCs at both mRNA and protein levels, with a period of about 1.5 h. Consistent with previous study, the peak value and period of Hes1 rhythm in human NPCs are not stable but vary from cycle to cycle and from cell to cell (Masamizu et al., 2006; Shimojo et al., 2008). The difficulties of obtaining human NPCs obstructed the measurement of Hes1 mRNA and protein level at multiple time points using the same generation of the same NPCs strain, which caused the slight incorrespondence between the shown representative results of Hes1 mRNA and protein rhythms (Figure 1A, 1B). The half-life of endogenous Hes1 protein in human NPCs is determined to be 17.6 ± 0.2 min, which is slightly shorter than that of the exogenously overexpressed Hes1 in mouse fibroblasts (Hirata et al., 2002).
Oscillatory Hes1 expression, as well as the Hes1 protein level, is essential to maintain the progenitor character of NPCs and thus plays a key role in neural development (Ohtsuka et al., 2001; Kageyama et al., 2008). Here, we report for the first time that HCMV infection disrupts rhythmic expression of the neural stem cell factor Hes1. Our previous work has shown that HCMV dysregulates Notch signaling of human NPCs by down-regulating at least two components, namely Notch1 including its intracellular domain (NICD) and the Notch1 ligand Jag1 (Li et al., 2015). As the downstream effector in the Notch signaling pathway, the expression of Hes1 was also positively regulated by the activation of upstream Notch signaling (Kageyama et al., 2008). But since the down-regulation of NICD1 and Jag1 initiated no early than 24 hpi of HCMV infection, and Hes1 expression dysregulation was observed as early as at 3 hpi, this Hes1 expression alteration at IE stage is irrelevant to the upstream Notch signaling change, but rather directed induced by HCMV infection. However, at later time points, the Hes1 expression alteration might be contributed by multiple influences, including both direct influence of HCMV infection and the indirect influence due to the HCMV induced upstream signaling dysregulation.
During development, NPCs undergo symmetric and asymmetric cell division to change their competency, giving rise to distinct cell types (Alvarez-Buylla et al., 2001; Fishell and Kriegstein, 2003; Gotz and Huttner, 2005). If these cells are prematurely depleted, not only the cell number is reduced but also the later-born cell types are lacking. So it is essential to maintain NPCs amount and stemness until the final stage in development, both in numbers and with a full spectrum of cell types (Hatakeyama et al., 2004; Kageyama et al., 2009). Our data demonstrated HCMV infection down-regulates Hes1 expression, and Hes1 depletion abnormally influence the NPCs characters, including the proliferation and neurospheres formation. This implies a potential mechanism of HCMV pathogenesis that viral infection leads less NPCs amount by repress self -renewal.
An oscillatory cycle of Hes1 expression is thought to be essential for maintaining the stem cell status of NPCs and governing their differentiation towards neurons or glia, which, in turn, controls fetal brain development (Ohtsuka et al., 2001; Kageyama et al., 2008). The four NPCs markers Sox2, DCX, GFAP and Nestin play critical roles in neural differentiation and/or maintenance of the multipotent state of the NPCs(Luo et al., 2010), and our data showed that depleting Hes1 in NPCs down-regulates Sox2 and DCX, and up-regulates GFAP and Nestin. DCX plays an important role in neuron migration signaling during brain development and is a marker of early migratory neuroblasts (Gleeson et al., 1998; Sossey-Alaoui et al., 1998). SOX2 is a transcription factor involved in embryonic development and stem cell self-renewal, SOX2 downregulation is associated with, and necessary for, both glial and neuronal differentiation(Episkopou, 2005; Suh et al., 2007; Luo et al., 2008). GFAP is first expressed in differentiating pluripotent stem cells and GFAP is used as a marker for differentiation of NPCs to an astroglial lineage (Luo et al., 2010; Pan et al., 2013). Nestin is an intermediate filament-forming protein that is downregulated during NPC differentiation (Lendahl et al., 1990; Sejersen and Lendahl, 1993). Thus, the Hes1 depletion induced NPCs marker alteration pattern doesn’t fit any normal differentiation direction, indicating that HCMV infection may drive NPCs abnormal differentiation toward a non-neuronal and non-glial lineage by dysregulating Hes1 expression, which powerfully disturbs the NPCs cell fate.
In summary, our study shows that oscillatory expression of Hes1, a critical regulator of Notch signaling and neural development, naturally exists in human NPCs. Hes1 rhythm is likely maintained by a negative feedback loop that depends on a short Hes1 t1/2 of < 18 min. HCMV infection potentially suppresses NPCs proliferation/neurosphere formation, and drives their abnormal deferrization via disrupts Hes1 rhythm and down-regulating its expression. This study reveals not only the expression rhythm of Hes1 in naïve human NPCs, but also suggests a novel mechanism for fetal brain development disorders induced by congenital HCMV infection.
-
This work was supported by the National Natural Science Foundation of China (31600145).
-
The authors declare that they have no conflict of interest. NPCs and human embryonic lung fibroblast cells (HELs) were isolated from postmortem fetal embryo tissues of brain or lung. The cell isolation procedures and research plans were approved by the Institutional Review Board (IRB) (WIVH10201202) according to the Guidelines for Biomedical Research Involving Human Subjects at Wuhan Institute of Virology, Chinese Academy of Sciences. The need for written or oral consents was waived by IRB.
-
MHL raised up the idea. MHL and FZ instructed the study. XJL and SNH performed the experiments. XJ provided NPCs samples. XJL, WBZ and MHL wrote the manuscript. All authors read and approved the final manuscript.
Human cytomegalovirus infection dysregulates neural progenitor cell fate by disrupting Hes1 rhythm and down-regulating its expression
- Received Date: 16 February 2017
- Accepted Date: 28 March 2017
- Published Date: 24 April 2017
Abstract: Human cytomegalovirus (HCMV) infection is a leading cause of birth defects,primarily affecting the central nervous system and causing its maldevelopment.As the essential downstream effector of Notch signaling pathway,Hes1,and its dynamic expression,plays an essential role on maintaining neural progenitor/stem cells (NPCs) cell fate and fetal brain development.In the present study,we reported the first observation of Hes1 oscillatory expression in human NPCs,with an approximately 1.5 hour periodicity and a Hes1 protein half-life of about 17(17.6±0.2) minutes.HCMV infection disrupts the Hes1 rhythm and down-regulates its expression.Furthermore,we discovered that depleting Hes1 protein disturbed NPCs cell fate by suppressing NPCs proliferation and neurosphere formation,and driving NPCs abnormal differentiation.These results suggested a novel mechanism linking disruption of Hes1 rhythm and down-regulation of Hes1 expression to neurodevelopmental disorders caused by congenital HCMV infection.














 DownLoad:
DownLoad: