-
Dear Editor,
As a major protein of cypovirus capsid shell, VP5 performs as the clamp protein to stabilize the capsid shell structure (Yu et al., 2008). And as a part of viral RNA (vRNA) replication machinery, VP5 has been found to possess an ATP-independent RNA chaperoning activity (Yang et al., 2014). Here, we report that VP5 also has the nucleoside triphosphatase (NTPase) activity, which can hydrolyze all kinds of NTPs and dNTPs. Thus, our finding reveals that VP5 is a multifunctional protein that may be involved in diverse processes during cypoviral life cycle.
The family Reoviridae is a large group of double-stranded RNA (dsRNA) viruses that include numerous important human and animal pathogens responsible for diseases in humans, livestock animals, insects and plants (Mertens, 2004). And the typical characteristic of dsRNA viruses is that their viral inner capsids are the machineries for vRNA replication. Reoviruses contain a layer of inner capsid shell and one or two outer layers of capsids. In this virus family, Rotavirus is the most famous genera, which infect vertebrates and is the main pathogenic virus that causes diarrhea and even death in children across the globe, while the genus cypovirus (also named cytoplasmic polyhedrosis virus, CPV) includes pathogens infecting insects. Different with other reoviruses and dsRNA viruses, CPVs only contain a single-layer capsid that is fully capable for vRNA replication. This unique structural feature makes CPV a simplified and ideal model for studying the replicative mechanism of vRNA replication of the Reoviridae or even all dsRNA viruses (Ahlquist, 2006). For example, the three-dimensional structure of CPV has been resolved and shows that the unique messenger RNA (mRNA) capping machinery is coupled with mRNA release hole (Yu et al., 2008). And the full atomic model of the CPV capsid shows how the nascent mRNA released from the hole and the mRNA capping progress during the releasing progress (Cheng et al., 2011; Yu et al., 2011).
The genomic RNAs of reoviruses and other dsRNA viruses are encapsidated into the inner core shell by a packing signal, then replicated by the RNA-dependent RNA polymerases (RdRPs) within the inner shell. The negative-stranded vRNAs [(–)-vRNAs] are retained in the core shell as replication templates and positive-stranded vRNAs [(+)-vRNAs] are exported from small holes of the shell to cytoplasm. The core shell ofcypovirus consists three main structural proteins, VP1, VP3, and VP5. Each CPV capsid contains 120 copies of VP1, 60 copies of VP3 and 120 copies of VP5. VP1 is the shell protein that forms the CPV capsid shell; VP3 is the mRNA-capping enzyme; and VP5 has been reported to be the clamp protein to stabilize the CPV capsid shell (Yang et al., 2012).
For reoviruses including CPVs, the 5′- and 3′-UTRs of reoviral (+)-RNA undergoes circularization to form a 5′-3′ panhandle-like structure, which is believed to play critical roles in encapsidation, translation, and vRNA replication by recruiting corresponding cellular or host machineries (Tortorici, 2006). On the other hand, this panhandle structure needs to be unfolded to make the 3′-end of vRNA accessible, thereby allowing the initiation of vRNA replication (Ahlquist, 2006). In our previous study, we has uncovered that the VP5 protein encoded by the type 5 Helicoverpa armigera cypovirus (HaCPV-5) has the RNA chaperoning activity that can unwind RNA helix and accelerate strand annealing independent of ATP. And CPV VP5 can act on the 5′-3′ panhandle structure of the CPV vRNA to destabilize the RNA structure and facilitate RNA replication in vitro (Yang et al., 2014). This is the first time to identify the RNA chaperoning activity associated with a dsRNA virus capsid protein.
As an RNA chaperone, the nucleic acid helix destabilizing activity of CPV VP5 does not require the hydrolysis of ATP or other NTPs to provide energy for melting basepairs. On the other hand, previous study also revealed that iflavirus nonstructural protein 2C, which is also an RNA chaperone, also possesses the NTPase activity (Cheng et al., 2013), although how the NTPase activity affects the activities of 2C remains unclear.
In order to further investigate the activity of CPV VP5, we expressed and purified MBP-fusion VP5 protein (MBP-VP5) using the Bac-to-Bac system as described previously (Cheng et al., 2013; Han et al., 2013). Standard procedures were used to construct the wild type plasmid of HaCPV5 VP5 and the mutations. All proteins were quantified by the Bradford method. Then we investigated the NTPase function of the VP5 using a direct colorimetric assay. A standard NTPase assay was carried out in a 20 μL reaction volume, containing 25 mmol/L HEPES-KOH (pH 8.0), 50 mmol/L NaCl, 1 mmol/L MgCl 2, 1 mmol/L DTT, 1 mmol/L NTP and 10 pmol of protein. Then the mixture was incubated at 37 °C for 20 min. 80 μL of dye solution (0.0812% malachite green, 5.72% ammonium molybdate, 2.32% polyvinyl alcohol and water were freshly mixed in a 2:1:1:2 ratio, respectively) was added into the reaction buffer and incubated at 37 °C for 5 min. Then 10 μL of 34% sodium citrate was added and the absorbance at 620 nm of the mixture was measured after 15 min. A standard curve of A620 nm versus known standard phosphate concentrations was used to determine the concentration of inorganic phosphate. All of the results given with this quantitative assay were averages of three separately repeated experiments.
To determine whether CPV VP5 has NTPase activity, we incubated purified MBP-VP5 with ATP, GTP, CTP, UTP, dATP, dGTP, dCTP or dTTP. The total amount of orthophosphate released after each reaction was measured to determine the NTPase activity of VP5. As shown in Figure 1A, VP5 could hydrolyze all four types of NTPs at different efficiencies. And the relative rates of hydrolysis were in the order ATP > GTP > UTP > CTP. Similarly, all four types of dNTPs could be hydrolyzed by VP5, and dATP was hydrolyzed most efficiently. Compared with their counterpart NTPs, each of the four dNTPs was more efficiently hydrolyzed ( Figure 1A). The data indicate that VP5 has NTPase activity that can hydrolyze all types of NTPs and dNTPs. Because ATP is the major energy source in cells (Hara, 2009), we chose ATP as the hydrolysis substrate in the subsequent assays.
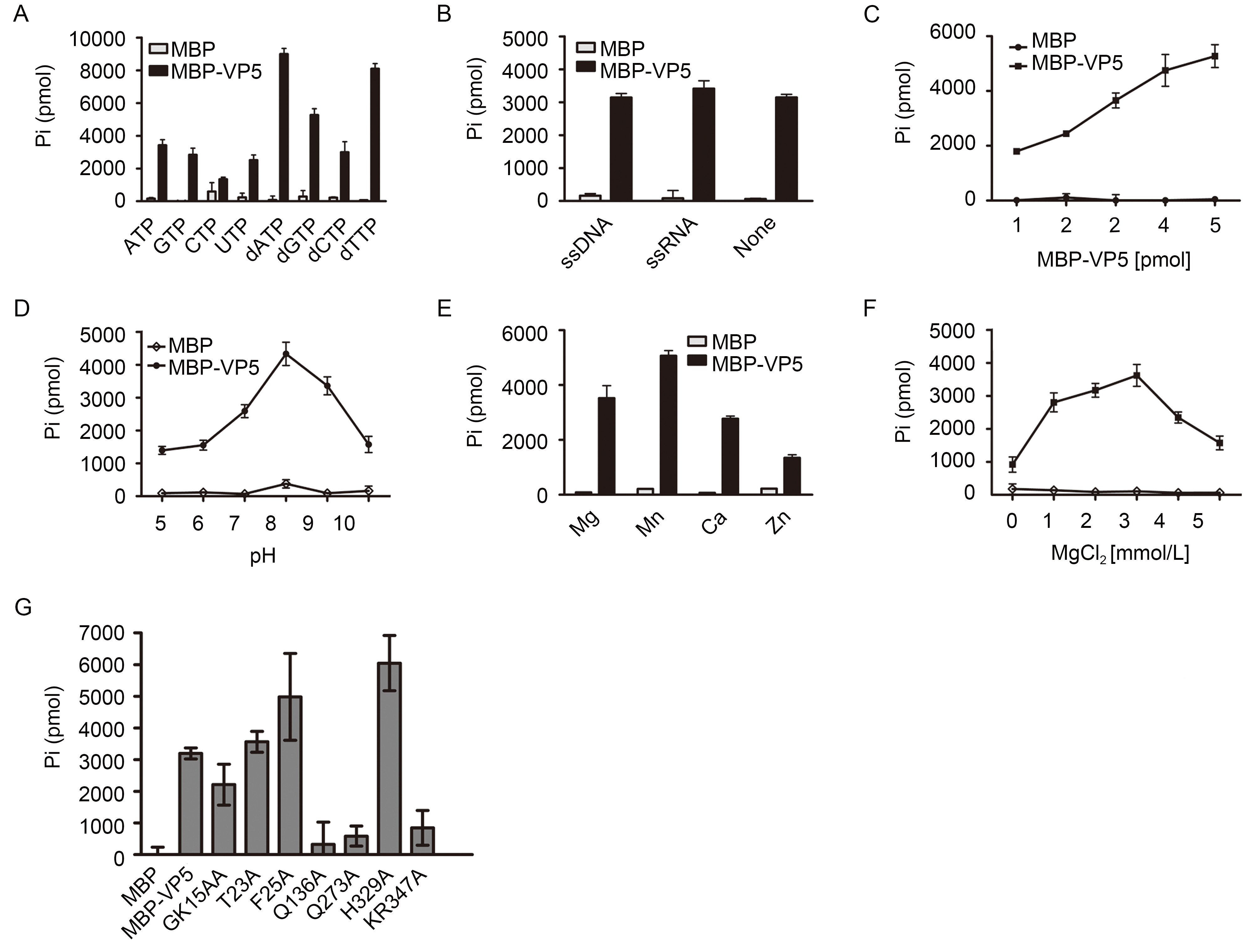
Figure 1. CPV VP5 has NTPase activity. (A) VP5 was reacted with the indicated NTP or dNTP. The ATPase activity of VP5 was determined with ssDNA or ssRNA (B), at the indicated concentrations of VP5 (C), at the indicated pH (D), with different divalent metal ions at the concentration of 2.5 mmol/L (E), or with indicated concentrations of MgCl2 (F). (G) The ATPase activity of WT or mutant VP5 protein as indicated was determined. (A-G) Error bars represent standard deviation (S.D.) values from three separate experiments
For many virus-encoded NTPases, the NTP hydrolysis activity can be stimulated by the presence of different kinds of nucleic acids (Incicco et al., 2013). Therefore, we added single-stranded DNA or RNA (ssDNA or ssRNA) to the reaction mixture to examine whether these nucleic acid strands would affect the efficiency of ATP hydrolysis. As shown in Figure 1B, the presence of ssDNA or ssRNA did not enhance the ATPase activity of VP5. In addition, the rate of enzyme activity usually changed with the protein concentration with defined reaction conditions (Cheng et al., 2013). Consistently, the increasing concentration of VP5 protein also gradually enhanced the ATP hydrolysis (Figure 1C).
After determining that CPV VP5 does have the NTPase activity, we also assessed the ATPase activity under varying conditions, such as pH and divalent metal ions, to further characterize its biochemical properties. To determine the optimum pH condition of VP5 NTPase activity, we conducted the ATPase assays at different pH values from 5.0 to 10.0. And our data show that CPV VP5 achieves its optimal ATPase activity at pH 8.0 (Figure 1D).
Most viral proteins that have NTPase activities require metallic ions for efficient NTP hydrolysis. To this end, we examined the requirement of different divalent metallic ions for VP5. Four divalent cations, Mg2+, Mn2+, Ca2+, and Zn2+, were supplemented into the reactions. As shown in Figure 1E, all these divalent ions supported the ATPase activity of VP5, and their efficiencies were as follows: Mn2+ > Mg 2+ > Ca 2+ > Zn 2+. Moreover, the VP5 showed the optimal ATPase activity in the presence of 3 mmol/L Mg2+, while higher concentrations of Mg2+ showed inhibitory effect to the ATPase activity of VP5 (Figure 1F).
Here, we found that CPV VP5 has both RNA helix destabilizing and ATPase activities. However, our previous study has revealed that the helix destabilizing activity is independent of ATP. It would be intriguing to examine if these two activities of CPV VP5 have any relations. To this end, a series of point mutations (GK16AA, T23A, F25A, Q136A, Q273A, H329A, and KR347AA) were constructed. These VP5 mutants were expressed and purified using Bac-to-Bac expression system as same as wild-type (WT) VP5. After that, the ATPase activity of WT or mutant VP5 was determined under the same condition. As shown in Figure 1G, the GK16AA, T23A, F25A, and H329A mutants of VP5 still showed the activity of ATP hydrolysis, while the Q136A, Q273A and KR347AA mutants of VP5 almost lost their ATPase activities. Our previous study has shown that the three mutants (T23A, F25A, Q273A) all lost their duplex destabilizing activity (Yang et al., 2014). But in this study, only one of them (Q273A) lost the ATPase while the other two (T23A, F25A) still possess the ATPase activity. The site Q273 may be very important for the whole protein structure. So when this site is mutated, the protein lost both of those two activities. Taken together, our results show that the RNA chaperoning and ATPase activities of HaCPV VP5 are not coupled.
It is interesting to ask why CPV VP5 has an ATPase activity that is not required or coupled with its known RNA chaperoning activity. Similarly, EoV 2C also possess both ATPase and ATP-independent RNA chaperoning activities (Cheng et al., 2013). For CPV VP5, it is probable that the ATPase activity involves in other activities, such as working as the clamp protein to stabilize CPV capsid. And in this scenario, the hydrolysis of ATP by VP5 might result in structural change of CPV capsid, possibly affecting CPV life cycle. This possibility will be examined in future studies. In conclusion, our study uncovered that HaCPV-5 VP5 has both RNA chaperoning and ATPase activities, and these two activities of VP5 are not coupled.
-
This work was supported by the National Natural Science Foundation of China grants No. 31400141 (to JY), the Natural Science Foundation of Hubei grant No. 2015CFB351 (to JY) and No. 2016CFA045 (to XZ), and the National Science Foundation for Post-doctoral Scientists of China grant No. 2015M572190 (to JY). The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Cypovirus capsid protein VP5 has nucleoside triphosphatase activity
- Received Date: 14 April 2017
- Accepted Date: 17 May 2017
- Published Date: 26 May 2017
Abstract: As a major protein of cypovirus capsid shell,VP5 performs as the clamp protein to stabilize the capsid shell structure (Yu et al.,2008).And as a part of viral RNA (vRNA) replication machinery,VP5 has been found to possess an ATP-independent RNA chaperoning activity (Yang et al.,2014).Here,we report that VP5 also has the nucleoside triphosphatase (NTPase) activity,which can hydrolyze all kinds of NTPs and dNTPs.Thus,our finding reveals that VP5 is a multifunctional protein that may be involved in diverse processes during cypoviral life cycle.














 DownLoad:
DownLoad: