-
Human cytomegalovirus (HCMV), called human herpesvirus 5, is a member of the human herpesvirus family, which also includes herpes simplex virus 1 (HSV-1), herpes simplex virus 2 (HSV-2), varicella zoster virus, Epstein-Barr virus, human herpesvirus 6, human herpesvirus 7, and Kaposi’s sarcoma-associated herpesvirus (Mocarski et al., 2013; Roizman et al., 2013). Like all other herpesviruses, HCMV contains a double-stranded DNA genome of more than 220 kb, which is packaged in an icosahedral shaped capsid. HCMV infectious particles, or virions, possess a lipid envelope that contains viral surface glycoproteins and encases the tegument compartment and the viral capsid (Liu and Zhou, 2007). The viral DNA genome serves as the basis for molecular detection of HCMV using nucleic acid-based amplification techniques such as polymerase chain reaction (PCR)-based methods (Barbi et al., 2000; Yamamoto et al., 2001; Boeckh et al., 2004; Yamamoto et al., 2006; Nozawa et al., 2007; Soetens et al., 2008; Boppana et al., 2010; Waggoner et al., 2012). Furthermore, viral capsid proteins and surface glycoproteins also serve as the antigens for the serological tests to detect previous exposure to HCMV and ongoing HCMV infection.
Like all other herpesviruses, HCMV can engage in lytic infection to produce viral progeny as well as establish latent infection that can co-exist with the host during the lifetime of the host (Mocarski et al., 2013; Roizman et al., 2013). HCMV is one of the most important opportunistic human pathogens. This virus usually causes asymptomatic infections in healthy or immunocompetent individuals but can lead to severe and potentially life-threatening complications in immune-immature individuals such as neonates or immune-compromised patients such as organ-transplant recipients and HIV-positive individuals (Britt, 1999). Disseminated infections such as HCMV-associated pneumonia, hepatitis, and retinitis were commonly found in AIDS patients before the advent of HAART therapy, while HCMV infection accounts for a majority of the deaths in organ transplant recipients. Even in immune-competent and healthy individuals, recent epidemiological evidences suggest that HCMV infection represents a significant risk for the development of cardiovascular diseases such as atherosclerosis and neoplasm such as human brain tumor and colon cancer (Mocarski et al., 2013).
While herpes simplex viruses (HSV) cause neonatal infections, HCMV is among the most common infectious agents to cross the human placenta and infect the developing fetus (Mocarski et al., 2013; Roizman et al., 2013). Indeed, HCMV infection is the leading congenital infection worldwide. In developed countries, HCMV is the leading non-genetic cause of sensorineural hearing loss in children (SNHL) (Demmler, 1991; Dahle et al., 2000; Morton and Nance, 2006; Goderis et al., 2016). Thus, HCMV infection represents a significant public health problem and causes substantial financial burden (Stratton et al., 2000). There are tremendous needs to generate rapid and accurate diagnostics of HCMV infection and develop effective approaches and compounds for the prevention and treatment of HCMV-associated diseases.
While several FDA-approved drugs have been shown to be effective in suppressing viral replication and lytic infection, they do not affect viral latent infection and therefore, cannot completely eliminate viral infection (Mocarski et al., 2013). Moreover, the use of these compounds has been associated with potential toxicity and side-effects. These issues, along with the emergence of drug-resistant viral strains, pose a need to develop new compounds and novel strategies for the prevention and treatment of HCMV infection. During the last several decades, substantial efforts have been made to develop both recombinant vaccines, inactivated virus vaccines, and live attenuated virus vaccines (Fu et al., 2014; Barry, 2015; Plachter, 2016). While HCMV vaccines that effectively prevent viral infections in vaccinated individuals are currently not available, several vaccine candidates have shown promising effects and are either in preclinical or clinical studies (Wang and Fu, 2014; Anderholm et al., 2016; Britt, 2017).
Efforts have been made to develop accurate and specific diagnostics for HCMV in the past. Several diagnostics have been approved by the FDA and are effective in detection of HCMV infection in adults and non-neonate populations (Britt, 1999). However, these methods are generally not applicable to large scale screening for detection of HCMV in neonates. In this review, we highlight the recent progress in diagnostic techniques that could potentially be used for the detection of HCMV infection in neonates and its direct implications in public health settings for diagnosing congenital HCMV infection.
-
HCMV infection is ubiquitous due to its highly efficient transmission primarily via casual contact (Boppana and Fowler, 2007; Cannon et al., 2010; Manicklal et al., 2013). In developed countries, HCMV sero-positivity in child bearing-age women ranges from less than 45% to 85%. In developing countries, the sero-positivity can reach almost 100%. Intrauterine HCMV transmission can occur via primary and non-primary infection (Boppana et al., 2001; Boppana and Fowler, 2007; Cannon et al., 2010; Manicklal et al., 2013; Marsico and Kimberlin, 2017). In primary infection, mothers are infected without preexisting immunity during pregnancy. Primary infection represents the highest risk for in-utero transmission with an infection rate of more than 30% (Boppana et al., 2001; Wang et al., 2011). In non-primary infection, HCMV replication and active infection can be found in women with preexisting immunity to HCMV. The in-utero transmission rate during non-primary infection is much lower (~less than 2%) than that in primary infection (Boppana et al., 2001; Pass et al., 2006; Belec and Brogan, 2011; Enders et al., 2011; Wang et al., 2011).
Congenital HCMV infection represents a significant public health problem (Kenneson and Cannon, 2007; Marsico and Kimberlin, 2017). In developed countries, its estimated incidence is around 0.5% of all live births and there are more than 60, 000 newborns with HCMV congenital infection annually in the US and the European Union alone. In developing countries, the estimated incidence of congenital HCMV infection is higher, ranging from 1% to 5% of all live births (Boppana et al., 2001; Pass et al., 2006; Enders et al., 2011; Wang et al., 2011; Marsico and Kimberlin, 2017).
Clinical manifestations of HCMV congenital infection range from asymptomatic infection to potentially life-threatening disease. For example, more than 85% of infected newborns are asymptomatic at birth (Kenneson and Cannon, 2007). Less than 15% exhibit clinical complications, which include hepatomegaly, splenomegaly, microcephaly, and other neurologic signs. Laboratory observations include chorioretinitis, thrombocytopenia, sensorineural hearing loss, and neuroimaging abnormalities associated with central nervous system (CNS) involvement (Kenneson and Cannon, 2007; Marsico and Kimberlin, 2017). However, the diagnostic criteria of symptomatic HCMV congenital infection is not well defined. In some instances, some cases consider subjects with low birth weight as symptomatic, whereas others do not (Boppana et al., 2001; Pass et al., 2006; Enders et al., 2011; Wang et al., 2011). In other instances, infants with abnormalities detected using specific testing such as SNHL, are considered as asymptomatic while others are not (Kenneson and Cannon, 2007).
Congenital HCMV infection is the leading viral cause of mental retardation (Kenneson and Cannon, 2007). Delays in neurodevelopment are common, and many affected infants suffer from cognitive disabilities, motor deficit, and visual impairment (Britt, 1999). Moreover, 50% of symptomatic and 10% of asymptomatic newborns develop some degree of hearing loss (Demmler, 1991; Dahle et al., 2000; Morton and Nance, 2006; Goderis et al., 2016). Among those who develop HCMV-related SNHL, complications of hearing loss may be present at birth or delayed in onset. Symptoms of late-onset SNHL may emerge within 3 years after birth and at least half of SNHL children exhibit further hearing deterioration as they age (Demmler, 1991; Dahle et al., 2000; Morton and Nance, 2006; Goderis et al., 2016). While there is currently no effective therapy offered to SNHL children, early identification and non-pharmacological interventions can reduce the hearing impairment and improve the language skills and psychological development of the young patients (Robinshaw, 1995; Yoshinaga-Itano, 1999).
Congenital HCMV infection presents a significant challenge to the public health system and a substantial economic burden to society. Many children with HCMV congenital infection demand constant and long lasting care, and need special interventional and educational services (Stratton et al., 2000). The annual cost associated with HCMV congenital infection in the US alone was estimated to be at least $2 billion in 1992 and would be much higher today (Stratton et al., 2000). Early diagnosis of congenital CMV infection will permit timely identification of newborns at risk of neurodevelopmental complications and will allow timely monitoring and appropriate intervention. Thus, there is a tremendous and urgent need to develop effective and practical diagnostics for screening neonates for HCMV congenital infection.
-
The high incidence of congenital HCMV infection and its significance as one of the most common causes of neurodevelopmental delay and SNHL in newborns promote the need for neonatal screening for HCMV infection. Currently, routine physical examination of newborns fails to identify more than 90% of children with HCMV congenital infection (Demmler, 1991; Dahle et al., 2000; Morton and Nance, 2006; Goderis et al., 2016). Moreover, complications associated with HCMV congenital infection, such as neurodevelopmental delays and SNHL, can develop after birth and may not be diagnosed by newborn screening for CMV-associated complications (Boppana et al., 2001; Pass et al., 2006; Belec and Brogan, 2011; Enders et al., 2011; Wang et al., 2011). Many HCMV-infected children may be asymptomatic at birth but their complications associated with HCMV infection progress during early childhood with late onset and progressive neurodevelopmental delay. Programs aimed to monitor, correct, and improve neurodevelopmental delay caused by HCMV infection demand early identification of infected newborns. These programs must begin early to prevent or reduce early-onset and late-onset complications. In the case of CMV-associated SNHL, the programs need to begin before crucial brain development for language and speech skills occurs (Robinshaw, 1995; Yoshinaga-Itano, 1999; Belec and Brogan, 2011). Furthermore, these programs need to focus on the molecular identification of HCMV infection itself rather than the complications associated with HCMV infection. However, neither antenatal nor neonatal HCMV screening programs are implemented in any countries today.
HCMV transmission among humans is highly efficient via casual contact and HCMV sero-prevalence increases in an age-dependent manner and proportionally with the progression of childhood (Kenneson and Cannon, 2007; Marsico and Kimberlin, 2017). Thus, screening for HCMV congenital infection can only be made with certainty at early days after birth, usually not more than two weeks after birth. After that, it will be difficult to distinguish congenital infection from postnatal infection. Numerous virological methods have been developed to diagnose prenatally acquired HCMV infection using various samples from neonates including saliva, urine, and dried-blood spot (DBS) (Barbi et al., 2000; Yamamoto et al., 2001; Yamamoto et al., 2006; Nozawa et al., 2007; Soetens et al., 2008; Boppana et al., 2010). However, their possible adaptation and applicable use for large-scale screening remain unproven.
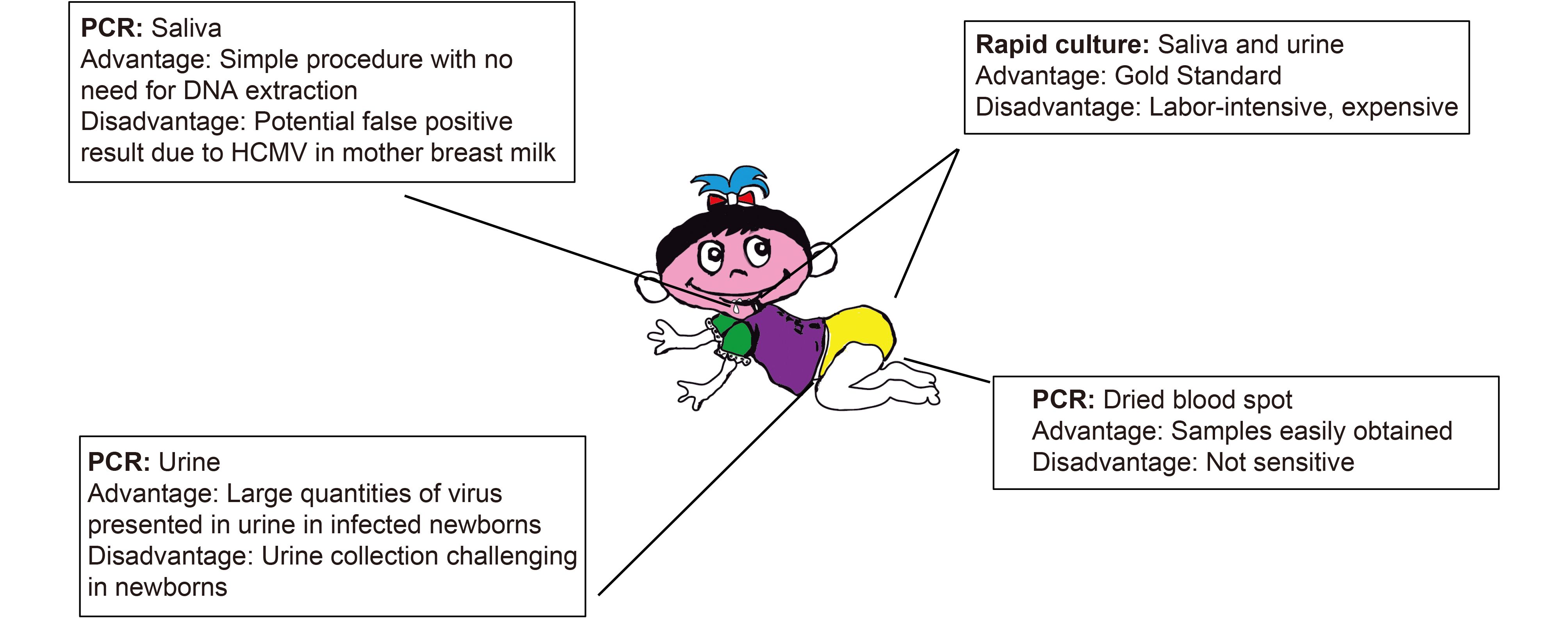
The current “gold standard” for the diagnosis of congenital HCMV infection in newborns is viral culture of urine or saliva samples (Figure 1) (Barbi et al., 2000; Yamamoto et al., 2001; Yamamoto et al., 2006; Nozawa et al., 2007; Soetens et al., 2008; Boppana et al., 2010). This assay is labor-intensive, expensive, and requires tissue culture facilities. Moreover, it is time-consuming and takes several days to complete. It is difficult to employ this assay for large scale screening in infants. Nucleic acid amplification techniques, such as PCR, are being used more frequently for the diagnosis of viral infection due to its great sensitivity, significant specificity, and quick turnaround (Sedlak and Jerome, 2013). During the last several years, there has been exciting progress on developing and using PCR-based methods for diagnosis of HCMV congenital infection (Figure 1) (Barbi et al., 2000; Yamamoto et al., 2001; Yamamoto et al., 2006; Nozawa et al., 2007; Soetens et al., 2008; Boppana et al., 2010; Boppana et al., 2011; Ross et al., 2014; Ross et al., 2015; Ross et al., 2017).
PCR and quantitative or real-time PCR (qPCR) have been widely used for diagnosis of viral diseases (Bustin, 2005). In qPCR reactions for diagnosis of viral infection, target viral DNA template is amplified with sequence-specific primers until it produces a signal through a sequence-specific fluorescent probe or a DNA intercalating dye. To detect lower levels of template DNA, more cycles of PCR are needed to generate a dye or fluorescent signal. The cycle of PCR amplification giving a signal is correlated with a known amount of DNA in a standard curve in order to determine the quantity of the DNA samples (Bustin, 2005).
Congenital HCMV infection fulfills the general criteria of the American College of Medical Genetics to be considered as a condition targeted for a newborn screening program. This is because congenital HCMV infection “can be identified as a time (within two days after birth) at which it would not ordinarily be detected clinically, and there are demonstrated benefits of early detection, timely intervention, and efficacious treatment of the condition” (Sweetman et al., 2006; Belec and Brogan, 2011). Screening and detection of HCMV in prenatal or early postnatal period has several benefits (Yow et al., 1988). First, early screening of congenital HCMV infection should aid timely identification of SNHL (Fowler et al., 1997). Furthermore, early screening may complement universal newborn hearing screening programs to identify neonates at risk for SNHL (Young et al., 2011). Correction of hearing loss with hearing aids or cochlear implants generates the best results when carried out before 6 months of age and greatly facilitates the development of normal language skills. Second, early detection of congenital HCMV infection should benefit timely intervention of HCMV-associated complications including neurodevelopmental delays. For example, a newborn positive for congenital HCMV should receive ophthalmologic examination to evaluate for CMV-associated retinitis. If any impairments are found, newborns with congenital HCMV may receive services to improve their cognitive, hearing, motor, speech, and vision functions.
-
Recent progress on the use of PCR-based assays for detection of congenital HCMV infection in newborns has generated much excitement in the public health field. A variety of methods have been evaluated for use in the diagnosis of congenital CMV infection (Barbi et al., 2000; Yamamoto et al., 2001; Yamamoto et al., 2006; Nozawa et al., 2007; Soetens et al., 2008; Boppana et al., 2010; Boppana et al., 2011; Ross et al., 2014; Ross et al., 2015; Ross et al., 2017). The most common and currently acceptable “gold standard” assay is culture-based testing of saliva, urine, and dried-blood-spot samples (Figure 1). PCR-based testing of dried-blood-spot samples have been evaluated because these samples are routinely collected in all infants, particularly in developed countries due to the implemented universal infant screening programs. However, results on the specificity and sensitivity of the PCR-based methods for testing are mixed, with some studies showing the methods as highly sensitive and specific while others suggesting the methods as not effective (Barbi et al., 2000; Yamamoto et al., 2001; Yamamoto et al., 2006; Nozawa et al., 2007; Soetens et al., 2008; Boppana et al., 2010; Boppana et al., 2011; Ross et al., 2014; Ross et al., 2015; Ross et al., 2017). The discrepancy of the results from these studies may partially be explained by the level of HCMV genome in the blood of the infants, especially in those who are asymptomatic, and the sensitivity of the PCR-based assays used. The study published by Boppana and colleagues in 2011 represents a milestone in developing promising techniques for newborn screening of congenital HCMV infection (Boppana et al., 2011). They employed PCR-based methods for testing both liquid and dried saliva samples of the newborns that were obtained at birth (Figure 1). Compared to the previous studies using PCR-based testing of dried-blood-spot samples, Boppana and colleagues took advantage of a very unique aspect of HCMV biology that HCMV engages in a high level of replication in the salivary gland and is constantly shed into saliva even in an asymptomatic individual (Boppana et al., 2011). Thus, saliva represents a better specimen for testing of CMV congenital infection.
In this landmark study, a total of 34, 989 infants born at seven medical centers in the US from June 2008 through November 2009 were enrolled (Boppana et al., 2011). Liquid saliva samples were collected in the phase 1 part of the study while dried saliva samples were collected in the phase 2 part. A quantitative PCR-based assay that was previously used for dried-blood-spot testing was used and the results from the PCR-based assay were directly compared to those from culture-testing experiments. A total of 177 out of 34, 989 infants (0.5%) were positive for HCMV, based on the three testing methods (culture testing of saliva, and PCR testing of liquid and dried saliva). The sensitivity and specificity of the liquid-saliva PCR assay were 100% and 99.9%, respectively. Moreover, the sensitivity and specificity of the dried-saliva PCR assay were 97.4% and 99.9%, respectively. Thus, quantitative PCR assays of both liquid- and dried-saliva samples exhibited high sensitivity and specificity for detecting CMV infection in newborns (Boppana et al., 2011).
The saliva rapid culture has been shown to have a sensitivity of at least 98% (Balcarek et al., 1993; Boppana et al., 2010). Although not perfect, rapid culture of saliva or urine samples at present remains the most commonly accepted standard method for testing of newborns for HCMV congenital infection. Quantitative PCR-based detection of HCMV in saliva has several advantages for HCMV screening in newborns (Figure 1) (Belec and Brogan, 2011; Boppana et al., 2011; Ross et al., 2014; Ross et al., 2015; Ross et al., 2017). First, saliva samples are easy to collect, even in infants during the first two days after birth. The collection can be easily carried out using non-invasive methods such as swabbing. Second, the collected dried saliva samples can be handled at room temperature with no need for refrigeration. The processing steps of these samples are expected to be easy and simple. Third, the procedure for PCR-based testing is simple and does not need a DNA extraction step. Furthermore, it does not require tissue culture facilities. Fourth, the PCR-based testing can be easily adapted for automation and for large-scale screening settings. Therefore, the PCR-based testing can be applied for newborn screening at relatively low cost. Fifth, PCR testing of saliva samples takes advantage of a unique aspect of HCMV biology that ongoing HCMV replication occurs in salivary glands and viruses are constantly shed into the saliva even in an asymptomatic individual. Thus, combined with the reasonable sensitivity and specificity of PCR, PCR-based testing of saliva samples is ideal for early detection of HCMV in infants, especially in those exhibiting no symptoms or complications associated with HCMV infection. Since the first report on PCR-based testing of saliva of infected infants, several follow-up studies had been carried out and recently reported (Boppana et al., 2011; Ross et al., 2014; Ross et al., 2015; Ross et al., 2017). These studies validated the results and conclusion from the original study by Bappopa and colleagues, and further demonstrated the utility and effectiveness of PCR-based testing of saliva for diagnosis and screening of HCMV in newborns.
Studies on PCR-based testing of saliva reveal several limitations of the methods for screening of congenital HCMV infections. First, a few samples had been shown to be false positives as these samples were identified as positive in both PCR-based assays of the liquid and dried saliva samples but negative in saliva rapid cultures when samples were collected within 1–2 days of birth. However, retesting of samples from these patients at a later day turned out to be negative. This can be partially explained as HCMV is occasionally shed in the breast milk of seropositive mothers and in the genital tract secretions of seropositive women at delivery (Kenneson and Cannon, 2007; Belec and Brogan, 2011; Marsico and Kimberlin, 2017). To avoid potential false positive results, a positive screening result should be verified by retesting the same individual within the first three weeks of age if saliva PCR is used to screen newborns. Second, saliva testing demands the collection of additional specimens (i.e. saliva) under the current universal newborn screening programs if large scale screening of saliva will be implemented. Third, the properties of PCR-based testing of saliva such as its sensitivity and specificity can be improved by the recent and continued advances in the nucleic acid-based amplification technology such as digital PCR approaches (Vogelstein and Kinzler, 1999; Bustin, 2005). Further studies on the PCR-based testing of saliva for diagnosis of HCMV congenital infection will address these issues.
-
PCR-based approaches have been routinely applied to be used for diagnosis of HCMV infection in immunocompromised hosts at risk for severe diseases, including AIDS patients and organ transplant recipients (Kenneson and Cannon, 2007). However, PCR-based methods have not been universally used for the diagnosis of HCMV congenital infection, and viral culture remains the accepted standard. It has been well known that infected individuals, including asymptomatic newborns, can shed large quantities of virus in saliva and urine, making these two specimens ideally suited for virus detection (Kenneson and Cannon, 2007). Traditionally, urine samples were used more frequently and could be collected using different methods. For example, urine specimens were collected using filter disks inserted into diapers of newborns (Nozawa et al., 2007; Inoue and Koyano, 2008). In more recent studies, urine specimens were collected using sterile bags and sterile cotton balls inserted into the diapers of newborns (Ross et al., 2014; Ross et al., 2015).
Until recently, few studies had reported on the comparison of urine and saliva specimens for the diagnosis of congenital HCMV infection (Figure 1). In 2006, Yamomoto and colleagues reported their study on 28 infants with congenital HCMV infection (Yamamoto et al., 2006). In this report, 24 cases (86%) were positive in the saliva PCR tests while 26 cases (93%) were positive in the urine PCR tests. During the last three years, several studies had been reported (Ross et al., 2014; Ross et al., 2015). In one study, newborns positive for HCMV by screening with either rapid culture or PCR were enrolled for follow-up evaluation, and as part of the confirmatory testing, saliva and urine samples were analyzed by both quantitative PCR and culture (Ross et al., 2014). This study directly compared the results of both PCR-based testing and rapid culture of saliva and urine specimens to determine if the quantitative PCR methods have the same clinical utility as the rapid culture methods for diagnosis of congenital HCMV infection (Ross et al., 2014). Moreover, this study also provided results to determine if the PCR-based methods perform equally well in both urine and saliva specimens. Eighty infants that had been identified to be positive for HCMV infection were enrolled in the follow-up study. Their saliva and urine samples were collected within the first three weeks of life (Ross et al., 2014).
The results of urine PCR and culture were positive in 79 (98.8%) and 76 (95%) of 80 urine samples, respectively (Ross et al., 2014). The results of saliva PCR and culture were positive in 80 (100%) and 78 (97.5%) of 80 saliva samples, respectively. When analyses were carried out to compare the data of PCR and culture of saliva and urine samples, the results of urine and saliva PCR were positive in 79 (98.8%) and 80 (100%) of 80 infants, respectively, while the results of urine and saliva culture were positive in 76 (95%) and 78 (97.5%) of 80 infants, respectively. These results show that PCR-based assays exhibited similar or better sensitivity and specificity than rapid culture of urine or saliva samples in the diagnosis of congenital HCMV infection (Ross et al., 2014).
In another report (Ross et al., 2015), 346 infants, who had been identified to be positive for HCMV infection either from rapid culture or PCR assays of saliva samples from 7 US medical centers, were enrolled in the follow-up study. Most of the urine samples were collected in sterile urine bags. At 3 of the 7 study sites, cotton balls were also used and placed in diapers to collect urines (Ross et al., 2015). This study was designed to compare the results of rapid culture and PCR assays when urine was collected by the traditional sterile bag method and by cotton balls. It appeared that the results of PCR assays were positive in 95% of urine samples regardless of the collection method. In contrast, the results of rapid culture were positive in much fewer urine samples collected using cotton balls (55.2%) than with samples collected in sterile urine bags (93.2%) from infants that were positive for HCMV in saliva. These results highlight the importance of the collection method for the urine specimens (Ross et al., 2015).
PCR-based testing of urine samples exhibited excellent performance in terms of sensitivity and specificity comparable to PCR-based testing of saliva samples and even better than the rapid culture of urine and saliva specimens (Belec and Brogan, 2011; Marsico and Kimberlin, 2017). However, this method still has several limitations. First, urine collection in newborns is very challenging. Collection using sterile urine bags is a complicated procedure in infants while samples collected using cotton balls in the diapers may not be as suitable for rapid culture testing as those collected in urine bags. It has been recently shown that urine samples that were collected on filter disks inserted in the diapers could be used for screening of congenital HCMV infection (Nozawa et al., 2007; Inoue and Koyano, 2008). However, urine collection from newborns still is not an easy task as the procedure requires collection steps that may be prone to potential contamination. Second, the performance of PCR-based testing of urine samples, with potentially more false positive and false negative results, was not better than that of PCR-based testing of saliva samples (Ross et al., 2014; Ross et al., 2015). Third, collection and processing of the urine samples requires additional steps and time that are not needed for saliva collection. Fourth, similar to PCR-based testing of saliva, the PCR-based testing of urine is difficult to adapt for the currently universal newborn screening programs which currently collect dried-blood-spot specimens.
-
Currently DBS samples are collected routinely for newborn metabolic and genetic screening from all infants born in developed countries in universal newborn screening programs (Belec and Brogan, 2011; Marsico and Kimberlin, 2017). Thus, it is reasonable to consider the possibility to use the collected DBS samples for diagnosis of congenital HCMV infection in infants (Figure 1). The collections of the DBS samples provide the sample basis to be used for large scale screening in newborns of congenital HCMV infection and alleviate the need for collection of additional samples in the universal newborn screening programs. Several previous studies reported that DBS PCR assays are highly sensitive in identifying infants with congenital HCMV infection and suggested that PCR-based testing of DBS specimens may represent a promising approach for HCMV screening in infants (Barbi et al., 2000; Revello and Gerna, 2002; Gohring et al., 2010; Leruez-Ville et al., 2011). However, more recent studies with larger numbers of clinical samples suggest otherwise. In a recent report, a multicenter study was designed on a scale comparable to that required for universal newborn screening (Boppana et al., 2010). The results of the PCR-based testing of DBS samples were compared to those of the culture of saliva specimens in 20, 446 newborns. These results showed that the PCR testing of DBS samples identified fewer than 40% of the HCMV infected infants (Boppana et al., 2010). Thus, the sensitivity of the PCR-based testing of DBS samples was unacceptably low in identifying newborns with congenital HCMV, compared to that of the “gold standard” rapid culture of saliva samples. In a follow-up study published in 2017 (Ross et al., 2017), whether the PCR-based testing of DBS samples can be used to identify infants with congenital CMV-associated SNHL was investigated. This study aimed to determine the ability of the DBS PCR assay for identification of infants with congenital HCMV at risk for disease and sequelae such as SNHL. Among the 313 infected newborns, 90 DBS samples (28.8%) were positive for CMV, with DBS PCR as positive in 9 of 28 (32.1%) symptomatic newborns and 81 of 285 (25.9%) asymptomatic newborns at birth (Ross et al., 2017). In order to study if DBS PCR can identify infants with HCMV-associated SNHL, the results of DBS PCR of infants with hearing loss at birth were compared to those with normal hearing. DBS was positive in 12 of 26 (46%) infants with SNHL at birth and 78 of 287 (27%) infants with normal hearing at birth. The PCR-based testing of DBS samples exhibited a sensitivity of 46.2% and a specificity of 72.8% in identifying newborns with SNHL at birth (Ross et al., 2017).
To further determine if the PCR-based testing of DBS samples can be used for identification of infants with late-onset SNHL, the results of DBS PCR assays with infants exhibiting SNHL at age 4 were compared to those from infants with normal hearing at the same age. These studies revealed that the results of PCR-based testing of DBS samples were positive in 11 of 26 (42%) of children with SNHL and 72 of 270 (27%) children with normal hearing at age 4 years, respectively (Ross et al., 2017). Of 277 asymptomatic individuals, DBS at birth was positive in 9 of 20 (45%) SNHL children by age 4 years and 70 of 257 (27%) with normal hearing by the same age, respectively. Of symptomatic individuals, DBS at birth was positive in 2 of 6 (33.3%) SNHL children by age 4 years and 2 of 12 (15.4%) with normal hearing by the same age, respectively. The PCR-based testing of DBS samples exhibited a sensitivity of 42.3% and a specificity of 73.3% in identifying children with SNHL at age 4 years. Further studies also showed that there was no difference in median DBS HCMV viral load between symptomatic and asymptomatic infants at birth and between SNHL children and those with normal hearing at age 4 (Ross et al., 2017). These observations demonstrated that PCR-based testing of DBS samples for HCMV DNA in newborns has low sensitivity and specificity for identification of infants with congenital HCMV infection and HCMV-associated SNHL at birth as well as at 4 years of age. Together, these recent studies suggest that PCR testing of DBS samples may not be suitable for the identification of infants infected congenitally and those at risk for HCMV-associated SNHL.
PCR-based testing of DBS samples offers several advantages and limitations (Belec and Brogan, 2011; Marsico and Kimberlin, 2017). One of the most important advantages is the existence of DBS specimen collection and the transportation procedures of DBS samples. DBS testing for congenital HCMV infection in newborns can be easily included in the current universal newborn screening program (Kharrazi et al., 2010). Thus, compared to testing of saliva and urine, DBS testing represents a significant advantage and can be more efficiently implemented when large-scale universal screening for congenital HCMV infection is considered as a part of the public health universal newborn screening programs for metabolic and genetic disorders. It is known that symptomatic infants have higher circulating HCMV viral loads in whole blood than asymptomatic infants (Lanari et al., 2006; Walter et al., 2008; Boppana et al., 2010; Leruez-Ville et al., 2011). Similarly, newborns with severe sequelae usually exhibit high level of viremia (Bradford et al., 2005). Thus, PCR-based testing of DBS samples may potentially be an efficient method for identification of infants at high risk of severe long-term sequelae (Britt, 1999; Leruez-Ville et al., 2011). However, significant technical hurdles needed to be overcome as recent studies suggest that less than 40% of cases of congenital HCMV infection and HCMV-associated SNHL can be identified by DBS PCR assays. These observations can be partially explained by the fact that symptomatic newborns may be infected many weeks before birth and have very low viral loads in blood during the perinatal period when testing is performed. The performance of the PCR-based DBS assays, such as sensitivity and specificity, can be improved by modifying the DNA extraction protocols, choosing the appropriate PCR technology (e.g. quantitative PCR vs digital PCR), and optimizing the PCR primer sets and the target HCMV DNA sequence. With the increased sensitivity and specificity of the PCR assays, DBS PCR testing may represent a promising and convenient method for large-scale screening of infants for congenital HCMV infection and HCMV-associated SNHL.
-
Recent studies on using PCR-based assays for identification of congenital HCMV infection have generated substantial enthusiasm and excitement to apply these approaches for screening newborns in order to detect HCMV infection early and prevent HCMV-associated complications effectively. Further studies may be needed to develop these approaches with the following considerations.
-
The qPCR-based assays have performed very well in diagnosis of viral infection in various specimens including saliva, urine, blood, and cerebrospinal fluid (Figure 1) (Bustin, 2005). However, they still have several limitations, including the need for a standard curve for the quantification of samples and the lack of universal standards of known quantity. These limitations significantly contribute to intra- and inter-laboratory imprecision and lack of commutability.
An innovative PCR-based method, called digital PCR (dPCR), has previously been developed and is being explored for detection of viral infections (Vogelstein and Kinzler, 1999). The dPCR technology uses the same primers and probes/dyes as qPCR but partitions a single bulk reaction into thousands or millions of separate microliter to picoliter scale reactions (Sykes et al., 1992; Vogelstein and Kinzler, 1999). Each reaction is cycled to the endpoint and each partition is read as positive or negative for target DNA. The number of positive partitions is analyzed to give a direct readout or absolute quantitation of the target DNA, eliminating the need to use standard curve in qPCR (Sykes et al., 1992; Vogelstein and Kinzler, 1999).
Previous studies have shown that dPCR is a promising method for diagnosis of viral diseases (Vogelstein and Kinzler, 1999; Baker, 2012; Sedlak and Jerome, 2013). The dPCR technology offers several advantages over qPCR, which currently represents the “gold standard” for molecular diagnosis of viral infections (Hindson et al., 2013; Sedlak and Jerome, 2013). First, dPCR exhibits precision superior to qPCR. Second, dPCR exhibits comparable sensitivity as qPCR. Third, dPCR can be used to standardize quantitation (Hindson et al., 2013; Sedlak and Jerome, 2013). Fourth, dPCR may be more resistant to components that are known to inhibit qPCR and regular PCR reactions because it has been reported that droplet dPCR was more refractory to inhibition than qPCR in the presence of heparin and SDS, common inhibitors in DNA extractions of clinical samples (Dingle et al., 2013). Fifth, dPCR is less affected by target sequence variability because it has been reported that droplet dPCR may be less susceptible than qPCR to strain mismatches in primer and probe sequences (Strain et al., 2013).
The dPCR technology has several limitations compared to qPCR. Although dPCR does not require a traditional standard curve, precisely and accurately defined calibrated materials are still needed to ensure commutability between clinical diagnostics laboratories. Compared to qPCR, dPCR is more expensive, and demands additional steps and more time to complete the assays (Hindson et al., 2013; Sedlak and Jerome, 2013). Many current dPCR assays are in an “open system” that is potentially more prone to contamination. Future efforts are needed to address these issues and improve the dPCR system in order to develop this innovative technology for high-throughput molecular diagnosis of viral infections.
-
Recent studies clearly demonstrated that choices of the types of tissues to be collected and the methods used for the collection play important roles in the screening results (Boppana et al., 2011; Ross et al., 2014; Ross et al., 2015; Ross et al., 2017). With the current PCR-based assays exhibiting excellent sensitivity and specificity, saliva specimens appear to be much more suitable for the PCR-based approaches for identification of congenital HCMV infection than the urine and DBS samples. Furthermore, saliva can be easily collected in newborns. However, unlike DBS samples, saliva is not a part of the specimens to be collected in the universal newborn screening programs that are currently implemented in developed countries including the US. With the increased sensitivity and specificity of the PCR assays including those by dPCR and conventional qPCR, DBS PCR testing may represent a promising and convenient method for large-scale screening of infants for congenital HCMV infection and HCMV-associated SNHL. Further studies on these issues will facilitate the development of the PCR-based approaches for the identification of congenital HCMV infection in newborns and the establishment of a universal newborn screening program for congenital HCMV infection.
-
We are grateful to Phong Trang, Hao Gong, Marco Paliza-Carre, Gia-Phong Vu, and Ting Wang for critical comments, insight discussions, and editorial assistance. This research has been supported by grants from Guangdong Innovative and Entrepreneurial Research Team Program (No. 2014ZT05S136), the National Mega Project on Major Infectious Disease Prevention (2012ZX10002006-003 and 2012ZX10004-207), and NIH (RO1-AI041927, RO1-AI091536, RO1-DE023935, and RO1-DE025462).
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Detection of congenital cytomegalovirus in newborns using nucleic acid amplification techniques and its public health implications
- Received Date: 02 August 2017
- Accepted Date: 23 October 2017
- Published Date: 30 October 2017
Abstract: Human cytomegalovirus (HCMV), a herpesvirus, is an important human pathogen that causes asymptomatic infections in healthy or immunocompetent individuals but can lead to severe and potentially life-threatening complications in immune-immature individuals such as neonates or immune-compromised patients such as organ-transplant recipients and HIV-positive individuals. Congenital HCMV infection represents a significant public health issue and poses substantial healthcare and economic burden to society. This virus causes the most common viral congenital infection worldwide, and is the leading non-genetic cause of sensorineural hearing loss in children in developed countries. Congenital HCMV infection is believed to fulfill the criteria of the American College of Medical Genetics to be considered as a condition targeted for a newborn screening program. This is because congenital HCMV infection can be identified during a time (within 2 days after birth) at which it would not ordinarily be detected clinically, and there are demonstrated benefits of early detection, timely intervention, and efficacious treatment of the condition. Recent progresses in developing polymerase chain reaction-based approaches to detect HCMV in samples obtained from newborns have generated much excitement in the field. In this review, we highlight the recent progress in diagnostic techniques that could potentially be used for the detection of HCMV infection in neonates and its direct implications in public health settings for diagnosing congenital HCMV infection.














 DownLoad:
DownLoad: