HTML
-
Gene therapy is a promising option for the treatment of hereditary and acquired diseases. Success in gene therapy requires a safe and effective gene delivery system that can provide and maintain an adequate level of gene expression. Retrovirus and lentivirus, members of Retroviridae family, have been extensively developed as gene delivery vectors. Lentiviral vectors have advantages for delivering genes to target cells. Compared to retroviral vectors, which can only transduce dividing cells due to pre-integration complex inability to cross the nuclear membrane, lentiviral vectors can transduce non-dividing cells (Naldini and Verma 2000; Okitsu et al. 2003). Lentiviral expression systems provide prolonged transgene expression (Varmus 1982). Also, lentiviral vectors are considered safer than gammaretroviral vectors in term of their reduced genotoxicity risk (Knight et al. 2012; Schambach et al. 2013). It is also known that gammaretroviral vectors have risks to the host due to potential oncogenesis caused by retroviral insertional activation of host genes (Varmus 1988). Here we determined if a lentiviral vector system could be optimized for the delivery of Nullbasic to Jurkat and primary CD4+ T cells. Nullbasic is a 101 amino acid mutant of the HIV-1 Tat protein that has anti-HIV-1 activity. It is based on the two-exon BH10 clone of Tat fused to the FLAG epitope, but differs from Tat where the basic domain was changed from the amino acid sequence RKKRRQRRR to GGGGGAGGG. As the basic domain is important for Tat:P-TEFb complex binding to TAR RNA and activation of the HIV-1 LTR promoter (Zhou et al. 2003; Kamori and Ueno 2017), Nullbasic does not activate HIV-1 transcription (Meredith et al. 2009). Nullbasic is considered to be a transdominant negative inhibitor of Tat because it can compete for binding with Tat to P-TEFb, a transcription factor essential for transactivation of the HIV-1 LTR promoter (Pearson et al. 1990; Ulich et al. 1996). However, Nullbasic also has unique abilities to inhibit additional steps of HIV-1 replication. Nullbasic interferes with HIV-1 replication by targeting Rev-mediated transport of HIV-1 mRNA (Lin et al. 2012) and reverse transcription (Meredith et al. 2009; Lin et al. 2015). These combined anti-HIV-1 activities permit strong inhibition of HIV-1 replication that makes Nullbasic a candidate HIV-1 inhibitor deliverable by retrovirus-based gene therapy vectors so as to prevent the spread of HIV infection in human T cells.
Using lentiviral vectors to transfer Nullbasic to target cells has been problematic. We previously demonstrated that specific cis-acting inhibition of VLP RNA reverse transcription by Nullbasic negatively effects transduction of target cells (Apolloni et al. 2013). Specifically, we used a lentiviral vector called pLOX, a second generation HIV-1-based lentiviral vector, to deliver a synthetic cDNA expressing Nullbasic fused to EGFP (NB-EGFP) or EGFP alone to Jurkat cells. However, the transduction rate of NBEGFP compared to EGFP using equivalent amounts of VLP was much lower for the former. NB-EGFP altered levels of RNA packaged into VLPs only slightly but greatly reduced levels of reverse transcription by NBEGFP VLPs when measured compared to control VLPs, which explained the reduced transduction efficiency. Inhibition of reverse transcription by Nullbasic has been investigated. A recent study showed that Nullbasic is packaged in virus particles, can bind to reverse transcriptase with a Kd of 40 nmol/L in vitro and virion cores isolated from Nullbasic-treated HIV-1 undergo accelerated disassembly compared to control HIV-1 cores (Lin et al. 2015). Defective reverse transcription is most likely the result of dysregulated core uncoating, as core uncoating and reverse transcription processes are closely linked (Hulme et al. 2011). Interestingly, wild type Tat has been shown to bind reverse transcriptase and improve HIV-1 reverse transcription efficiency (Apolloni et al. 2007), suggesting that Nullbasic may act as a transdominant negative inhibitor of reverse transcription. Given that pLOX and other HIV-1-based lentiviral vectors use the same viral apparatus for their early replication, the mechanism of Nullbasic inhibition of lentiviral vector VLP reverse transcription is likely to be similar.
Nullbasic can also inhibit the transport of HIV-1 unspliced and singly spiced mRNA from the nucleus to cytoplasm. Recently, it was shown that Nullbasic specifically targeted the cellular RNA helicase DDX1, which is a Rev binding protein that regulates its function in mRNA transport. DDX1 directly interacts with the multimerization domain of HIV-1 Rev protein to promote Rev oligomerization on the RRE (Fang et al. 2004; Robertson-Anderson et al. 2011). Interestingly, Nullbasic inhibition of Rev could be reversed by overexpression of DDX1-HA or TatFLAG (Lin et al. 2014).
Here, we hypothesized that overexpression of a Tat binding intrabody, or either Tat-FLAG, Rev or DDX1-HA could improve transduction efficiency of Nullbasic-treated VLPs by overcoming the blocks imposed by Nullbasic to virus replication.
-
Human embryonic kidney (HEK) 293T cells (ATCC® CRL-3216) were grown in Dubelcco's modified Eagle's medium (DMEM; Life Technologies, Carslbad, CA, USA) supplemented with 10% (v/v) FBS, penicillin (100 IU/mL) and streptomycin (100 lg/mL) (referred to as DF10 medium). Jurkat cells were grown in RPMI medium (Life Technologies) supplemented with 10% (v/v) FBS, penicillin (100 IU/mL) and streptomycin (100 lg/mL) (referred to as RF10 medium). HEK 293T-mCh-huTat2 and HEK293TmCh cell lines were established by transduction of pSicoREf1a-mCh-huTat2 and pSicoR-Ef1a-mCh-puro lentiviral systems respectively, and were then selected using FACS for the top 10% of mCherry expressing cells by mean fluorescent intensity.
-
Human buffy coats were supplied by Australian Red Cross Blood service (negative for HIV-1, hepatitis B and hepatitis C). PBMCs were isolated by Ficoll density gradient centrifugation of the buffy coat cells. CD4+ cells were isolated from the PBMCs by using a MACS human CD4+ cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. The selected CD4+ cells were grown in 6 cm tissue culture dishes and stimulated using plates pre-coated with purified anti-human CD3 (clone HIT3a) and anti-human CD28 (clone CD28.2) antibodies (BioLegend, San Diego, CA, USA) in RPMI medium supplemented with 20% (v/v) FBS and 5 ng/mL IL-2 (hereafter called RF20 IL-2) for 2 days. All cells were grown at 37 ℃ in humidified incubators with 5% CO2.
-
A plasmid containing the huTat2 intrabody cDNA was kindly provided by Wayne A. Marasco. A lentiviral expression plasmid pSicoR-Ef1a-mCh-Puro was a gift from Bruce Conklin (Addgene plasmid # 31845). Briefly, the gene encoding hu-Tat2 was fused at the C terminal of mCherry-T2A in the pSicoR lentiviral vector. A huTat2 with C terminal HA tag gene, huTat2-HA, was inserted into a pcDNA3.1+ plasmid. A pSicoR vector lentiviral plasmids expressing a Nullbasic-ZsGreen1 (NB-ZSG1) fusion protein or ZsGreen1 (ZSG1) were described previously (Jin et al. 2016). pNL4-3.Luc.R-E- was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH (Abbud et al. 1995; Connor et al. 1995; He et al. 1995). Tat101-FLAG, DDX1-HA and Rev expression plasmids were described previously (Lin et al. 2014).
-
VSV-G-pseudotyped HIV-1 harboring the firefly luciferase reporter gene was produced in HEK 293T cells by cotransfection of pNL4-3.Luc.R-E- and pCMV-VSV-G plasmids for 48 h. The supernatant was collected and CAp24 concentration was measured by ELISA (Zeptometrix, Buffalo, NY, USA), as recommended by the manufacturer. NB-ZSG1 and ZSG1 VLPs were produced in HEK 293T cells expressing mCherry-T2A-huTat2 or mCherry alone or parental HEK 293T cells in a 10 cm culture disk by co-transfection of 2 μg of pSicoR-Flag-NBZSG1 or pSicoR-ZSG1 plasmids, 2 μg of pCMV-VSV-G expressing plasmid and 6 lg of pCMVDR8.91 plasmid using 10 μL of X-tremeGENETM plasmid DNA transfection reagent (Sigma Aldrich, Castle Hill, NSW, Australia) according to the manufacturer's instructions. NB-ZSG1 and ZSG1 VLPs were also produced in HEK 293T cells where Tat, DDX1 and Rev were overexpressed alone or in combination by adding 3 μg of each plasmid in the transfected solution. Six hours post transfection, the cells were washed with PBS and the media was replaced. The VLPs were collected at 48 h post transfection and filtered through a 0.45 μm filter. CAp24 concentration was measured by ELISA (Zeptometrix), as recommended by the manufacturer.
-
Cell lysates were made from the HEK 293T expressing mCherry-T2A-huTat2 or mCherry or parental HEK 293T cells in cell lysis buffer (50 mmol/L Tris HCl pH 7.4, 150 mmol/L NaCl, 1 mmol/L EDTA and 1% (v/v) Triton X-100). The total protein concentration was measured by a Bradford assay using Bio-Rad protein assay (Bio-Rad, Hercules, CA, United States) and equivalent amounts of protein were used for analysis. To check mCherry-T2AhuTat2 expression in the cell lines, the blot was stained with anti-mCherry rabbit antibody (at 1:2000) (BioVision, Milpitas, CA, USA) followed by anti-rabbit IgG horseradish peroxidase (HRP)-linked antibody (at 1:2000) (Cell Signaling Technology, Danvers, MA, USA) and b-tubulin antibody (at 1:2000) (Sigma Aldrich) followed by antimouse IgG HRP-linked antibody (at 1:2000) (Cell Signaling Technology). The expression of Nullbasic, Tat, Rev and DDX1 was confirmed by staining the blot with rabbit anti-FLAG (at 1:2000) (Sigma Aldrich), mouse anti-Rev (at 1:500) (Santa Cruz Biotechnology, Dallas, TX, United States) and rabbit anti-DDX1 (at 1:500) antibodies (Santa Cruz Biotechnology) respectively.
-
Transduction was performed in 24 well culture dishes using 4 × 105 Jurkat or primary CD4+ T cells in each well. For each well, capsid amounts equal to 50, 10 or 2.5 ng of VLPs plus 8 ng/mL polybrene were added to the cells. Spinoculation was performed by centrifugation at 1500×g for 1 h at 32 ℃. The medium was replaced the next day. Samples were taken at 72 h post transduction, fixed by 1% paraformaldehyde in PBS, and then analyzed using a BD LSR 4 flow cytometer. Data were analyzed by version 9 FlowJo single cell analysis software.
-
Statistical analyses were performed using Student's t test or ANOVA and Tukey's multiple comparisons test on the data from at least three independent experiments or measurements where P values are shown. A confidence interval of 95% was used, therefore P value less than 0.05 were considered to be significant.
Cell lines and Cultures
Isolation of Primary CD4+ Cells
Generation of Plasmid Constructs
HIV-1 and VLP Production
Western Blot Analysis
Transduction of Jurkat Cells and Primary CD4+ T Cells by NB-ZSG1 or ZSG1 VLPs
Statistical Analysis
-
We tested the single chain variable region fragment humanized (hu) Tat2 intrabody (huTat2), which is a Tat antagonist agent (Mhashilkar et al. 1995b; Mhashilkar et al. 1999), to see if its overexpression could improve transduction by NB-ZSG1 VLPs. An intrabody is an intracellularly expressed recombinant antibody fragment that can bind a target protein within the cell. Earlier studies have shown that when delivered to human immune cells with gamma-retroviral gene therapy vectors, the huTat2 intrabody can protect cells from HIV-1 infection, replication and pathogenesis (Marasco et al. 1999; Braun et al. 2012; Kang et al. 2014). The huTat2 intrabody binds to amino acids 1–20 in the N-terminal proline-rich region of Tat (Marasco et al. 1999), which is entirely conserved in the Nullbasic protein. The gene encoding huTat2 was ligated into the lentiviral vector pSicoR-Ef1a-mCh-puro where huTat2 replaced the pac gene encoding puromycin N-acetyl-transferase (PAC) (Fig. 1). In this vector, the expression of mCherry is followed by a T2A ribosomal skipping sequence to allow for independent translation of huTat2. As no anti-huTat2 antibody is available for detection by western blot, detection of mCherry in cells acts as a surrogate marker for expression of huTat2. A stop codon inserted in the T2A sequence (Ryan et al. 1991) prevented translation of PAC in the control vector.
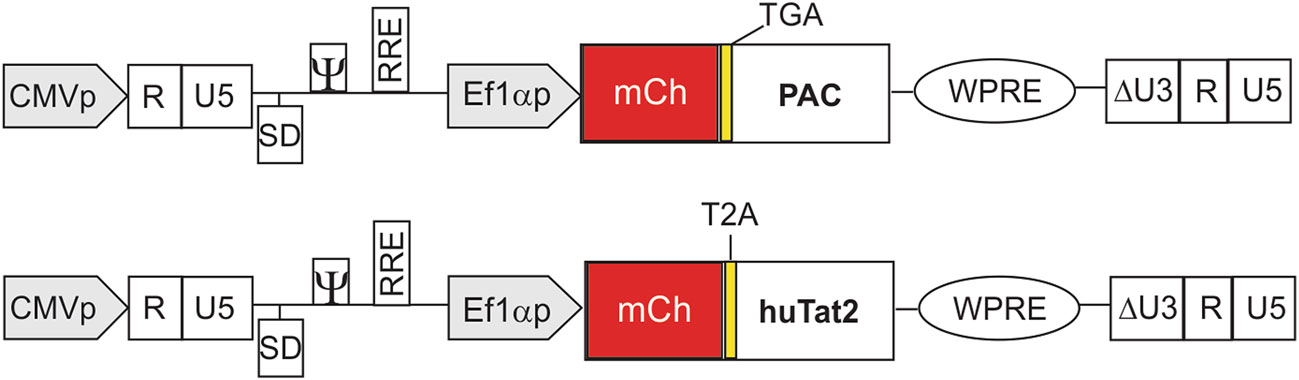
Figure 1. Schematic diagram of pSicoR-Ef1a-mCh-puro with PAC or huTat2 intrabody. The gene of interest is driven by Ef1α promoter. The gene encoding huTat2 was ligated into the lentiviral vector pSicoR-Ef1a-mCh-puro where huTat2 replaced the pac gene encoding puromycin N-acetyl-transferase (PAC). This transfer vector has a central polypurine tract (cPPT) (not shown) for assisting reverse transcription and gene translocation into the nucleus of non-dividing cells, RRE for assisting Rev function in transporting mRNA from the nucleus to cytoplasm, and a psi (w) packaging signal for viral genome packaging. For safety, this vector has an LTR U3 region deletion, creating a self-inactivating (SIN) (DU3) virus that prevents postintegration transcription of the full length viral genome from the LTR. A woodchuck hepatitis virus post-transcriptional modification element (WPRE) in the vector enhances the levels of transgene transcripts in the nucleus and cytoplasm of target cells (Zufferey et al. 1999; Bauer and Anderson 2014). The ribosomal skipping element T2A (Ryan et al. 1991) is located between mCherry and huTat2, which allows for independent translation of huTat2. A TGA stop codon was inserted into the control vector to prevent expression of PAC. In this vector, the expression of mCherry acts as a surrogate marker for huTat2 expression
VLPs were produced using each vector and used to transduce HEK293T cells and visualized by fluorescent microscopy (Fig. 2A). Cells that were mCherry positive were collected by FACS and the purity was confirmed by flow cytometry (Fig. 2B). Cell lysates prepared from the selected cells were analyzed by western blot using an antimCherry antibody. The mCherry-T2A protein was clearly detectable (Fig. 2C, middle panel), indicating that huTat2 was made. Because T2A ribosomal skipping is not completely efficient, a low amount of the * 65 kDa mCherryT2A-huTat2 fusion protein was observed following longer exposure of the western blot (Fig. 2C, top panel).
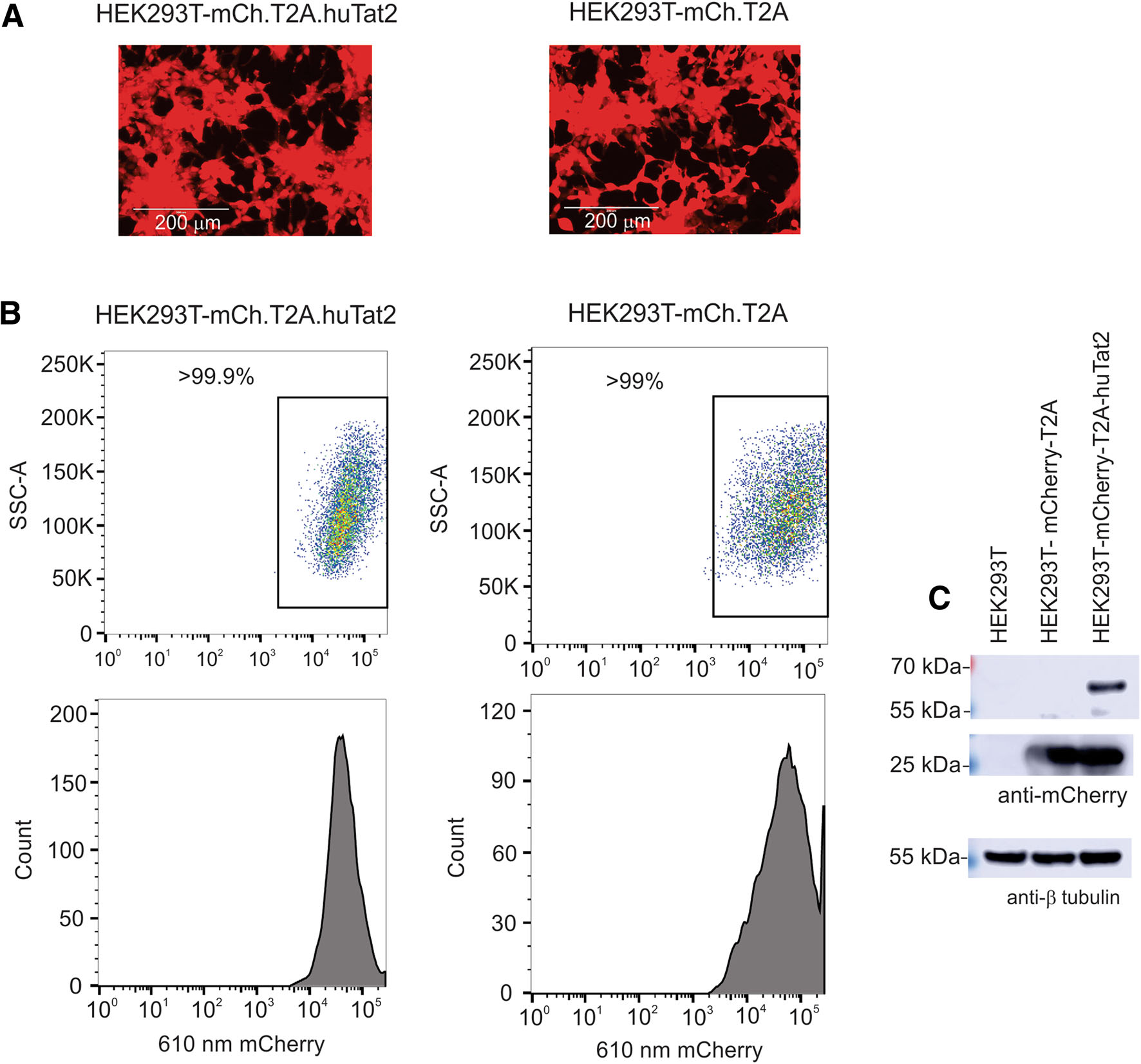
Figure 2. Expression of mCherry in HEK 293T cells. A HEK293T cells transduced with pSicoR-Ef1a-mCh-T2A-huTat2 or pSicoR-Ef1amCh-T2A expressed mCherry visualized using the EVOS imaging system. B The highly expressed mCherry cells were sorted by FACS and analyzed by flow cytometry. C mCherry expression in mCh-T2AhuTat2 and mCh-T2A cell lines was detected by western blot
The stable cell line HEK293T-mCh-huTat2 and HEK293T-mCh-T2A were infected with HIV-1 NL4-3.Luc.R-E- (Connor et al. 1995; He et al. 1995) pseudotyped with VSV-G envelope. This recombinant HIV-1 reporter virus has firefly luciferase inserted in the nef open reading frame and requires Tat for optimal expression of luciferase. After a 24 h infection, cell lysates were prepared and the level of firefly luciferase was measured. The results showed a statistically significant ~ 25–30% decrease in firefly luciferase activity in HEK293T-mChhuTat2 intrabody cells compared to control cells (Fig. 3A). The data indicate that the huTat2 intrabody can inhibit Tat transactivation of the HIV-1 LTR in HEK 293T cells, as previously reported by others using different cell types (Mhashilkar et al. 1995a).
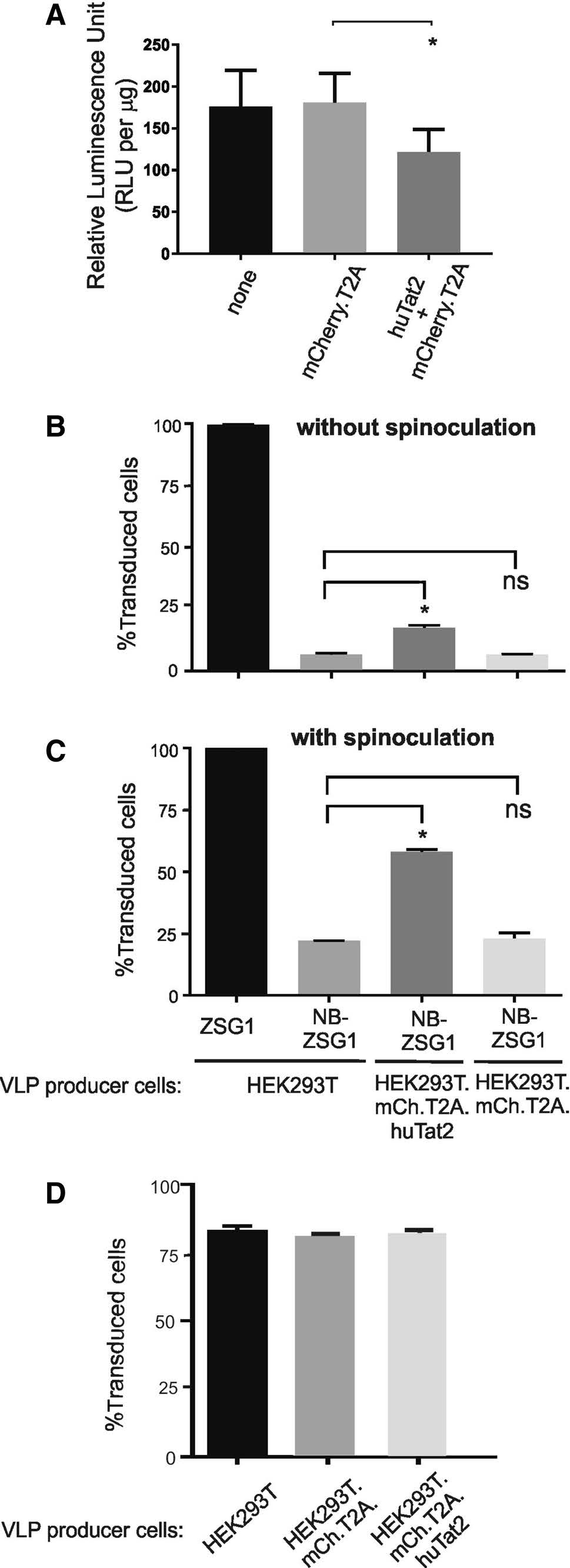
Figure 3. The huTat2 intrabody inhibits Tat-mediated transactivation and improves Nullbasic-VLP transduction of Jurkat cells. A HEK293T-mCh-huTat2 and HEK293T-mCh or parental HEK 293T cells were infected with VSV-G pseudotype HIV-1 luciferase reporter virus. B Jurkat cells were transduced with ZSG1 or NBZSG1 VLPs (equivalent to 50 ng CA) produced in VLP producer cell lines as indicated. Transduction was performed using polybrene without spinoculation; or C with spinoculation. D Jurkat cells were transduced with ZSG1 VLPs (equivalent to 10 ng CA) made by VLP producer cell lines as indicated. The transductions were performed using polybrene without spinoculation. All experiments were performed three times independently in triplicate. Bars indicate mean percent of transduced cells and error bars indicate the standard deviation. A asterisk designates a P value less than 0.05 and a ns designates not significant
Next we produced lentiviral-based VLPs that conveyed NB-ZSG1 or ZSG1 (see Jin et al. 2016) in HEK293TmCh-huTat2 and HEK293T-mCh-T2A cell lines to determine if huTat2 could help mitigate the negative effects Nullbasic has on lentiviral VLP transduction. Jurkat cells were transduced with equivalent amounts of each VLP normalized to total HIV capsid (CA), with or without spinoculation. Spinoculation can improve rates of infection by retroviruses and transduction efficiency of VLPs (Forestell et al. 1996). NB-ZSG1 VLPs suffer a defective reverse transcription due to NB-ZSG1 that is packaged in the VLP and so are less efficient at transduction than ZSG1 VLP (Apolloni et al. 2013). For these experiments, a VLP amount equivalent to 50 ng CA was selected for transduction experiments that permitted measurable and reliable transduction of Jurkat cells by VLP conveying NB-ZSG1. The cells were transduced and then incubated for 72 h, then the percentage of cells that were positive for ZSG1 or NBZSG1 was measured by flow cytometry. The results showed that[99% of Jurkat cells were successfully transduced by ZSG1-VLPs irrespective of spinoculation (Fig. 3B, C). This indicates that the concentration of ZSG1-VLPs used was saturating for the number of cells and the conditions used. However, at the same concentration of CA, 12% of Jurkat cells were successfully transduced by NB-ZSG1-VLPs without spinoculation (Fig. 3B), which increased to 25% with spinoculation (Fig. 3C). The results show that huTat2 improved transduction of Jurkat cells with NB-ZSG1-VLPs by ~ twofold when the VLPs were made in HEK293T-mCh-T2A-huTat2 cells compared to NB-ZSG1-VLPs made in HEK293T-mCh-T2A cells. We tested the effects of huTat2 on ZSG1-VLP as well (Fig. 3D). We reduced the amount of VLP used here by fivefold so that transductions rates were below 100%. Hence any positive or negative effects of huTat2 on ZSG1-VLP transduction rates could be detected. The transduction rate of Jurkat cells by control ZSG1-VLPs was unchanged irrespective of the HEK 293T cell line used, HEK293TmCh-T2A-huTat2 or HEK293T-mCh-T2A, to make the VLPs (Fig. 3D). Therefore, we conclude that huTat2 intrabody can specifically improve transduction rates of Jurkat cells by NB-ZSG1-VLPs, suggesting that huTat2 at least partially blocks Nullbasic inhibition of reverse transcription.
-
NB-ZSG1 VLPs and ZSG1 VLPs were produced in HEK293T cells by co-transfection of pSicoR-Flag-NBZSG1 or pSicoR-ZSG1 plasmids, pCMV-VSV-G plasmid and pCMVDR8.91 plasmid along with an empty pCDNA3.1+ plasmid or with plasmids encoding TatFLAG, Rev or DDX1-HA. Western blots were performed using lysates prepared from the transfected HEK293T cells to confirm expression of the gene products. The expression of NB-ZSG1, Tat-FLAG, DDX1-HA and Rev in the VLP producer cells was detected by western blot (Fig. 4A). The collected VLPs were used to transduce Jurkat cells with or without a spinoculation step using an equivalent amount of VLPs normalized to total CA in the supernatant. Irrespective of spinoculation, ZSG1-VLPs transduced > 99.9% of Jurkat cells under the conditions tested (Fig. 4B, C). However, the transduction rate of NB-ZSG1-VLP was ~ 25% without spinoculation and ~ 50% with spinoculation, respectively.
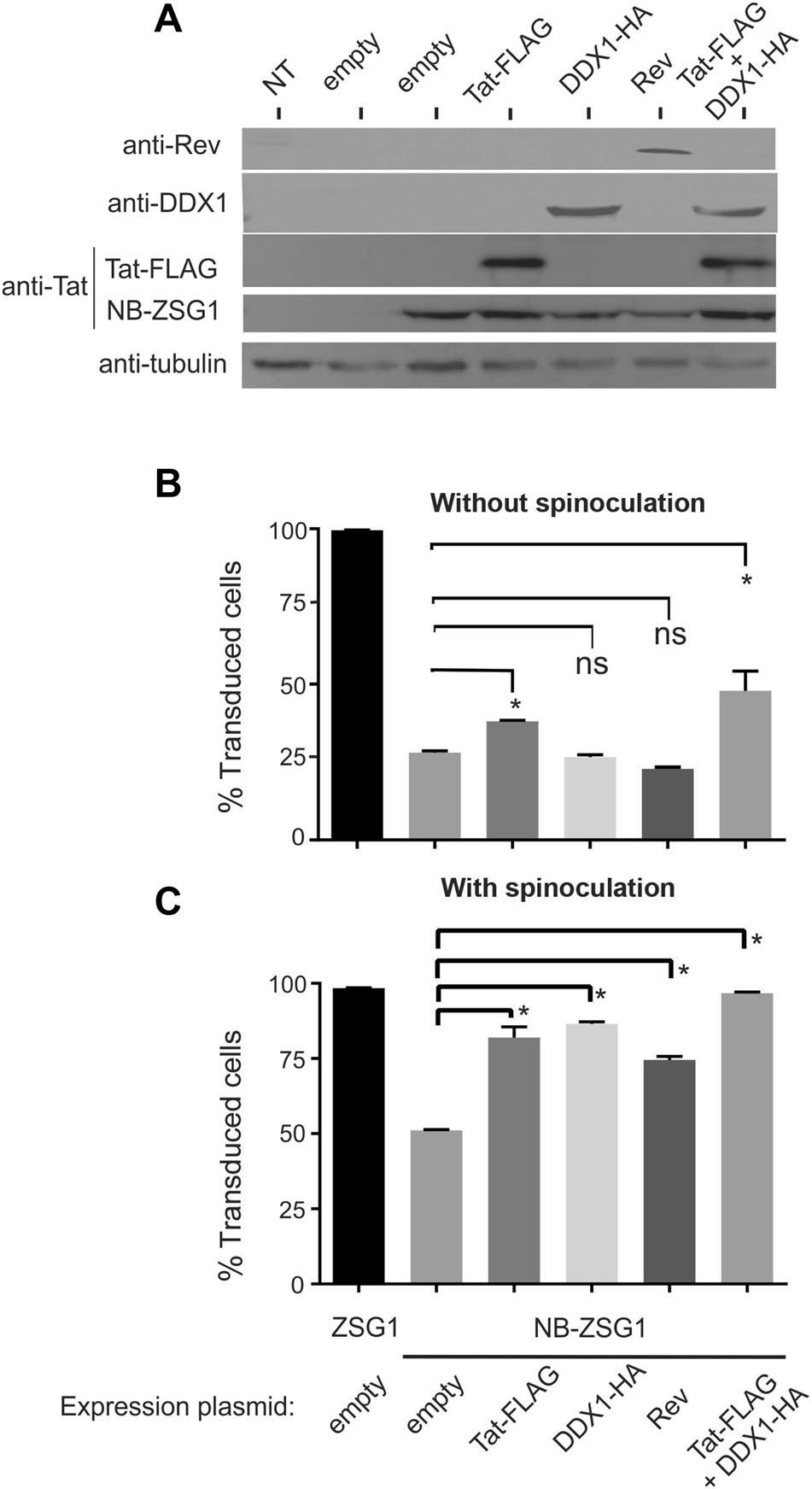
Figure 4. Transduction efficiency of Jurkat cells by NB-ZSG1 VLPs can be improved by overexpression of Nullbasic antagonists. A Detection of NB-ZSG1 and overexpressed Tat-FLAG, DDX1-HA and Rev by western blot. Lysates were collected from the VLP producer cells and stained with anti-FLAG (to detect both Tat-FLAG and NB-ZSG1), anti-Rev, anti-DDX1 and anti b tubulin antibodies. B VLPs equivalent to 50 ng of CA, produced in HEK 293T by co-transfection of VLP producer plasmids and Tat, Rev and/or DDX1 (as indicated), were used to transduce Jurkat cells without spinoculation. C The same VLPs as in B were used to transduce Jurkat cells using spinoculation. All experiments were performed three times independently in triplicate. Bars indicate mean percent of transduced cells. Bars indicate mean percent of transduced cells and error bars indicate standard deviation. A asterisk designates a P value less than 0.05 and a ns designates not significant
Without spinoculation, the transduction rate of NBZSG1 VLPs was improved when Tat-FLAG was overexpressed in the VLP producer cells (Fig. 4B), but not when DDX1-HA or Rev were overexpressed. With spinoculation, Tat-FLAG, Rev or DDX1-HA overexpression improved the transduction rate of NB-ZSG1 by ~ 75%–90%. Co-expression of Tat-FLAG and DDX1-HA improved transduction; this was most apparent when spinoculation was used, where > 95% of Jurkat cells expressed NB-ZSG1 (Fig. 4C). Next, the assays were repeated using reduced amounts of VLP so that the level of transduction by ZSG1-VLP was non-saturating (Fig. 5). In this experiment, ZSG1-VLP transduced ~ 25% (Fig. 5A) and ~ 60% (Fig. 5B) of Jurkat cells without and with spinoculation, respectively. Nevertheless, NB-ZSG1 transduction was improved when the VLP producer cells were supplied each NB antagonist, although improved transduction was more apparent when spinoculation was used (Fig. 5B), where all increased transduction rates observed were statistically significant. VLP produced in cells provided Tat-FLAG or Tat-FLAG and DDX1-HA demonstrated approximately twofold higher transduction rates compared to cells treated with empty vector. B: A small improvement in transduction rates were observed for VLP made by producer cells treated with DDX1-HA or Rev.

Figure 5. Improved rates of Jurkat cell transduction observed using a reduced amount ofNB-ZSG1 VLP. The transductionof Jurkat cells with (A) and without spinoculation (B) was repeated as in Fig. 4 using 2.5 ng CA of ZSG1-VLP or NB-ZSG1-VLP produced in HEK 293T cells treated as indicated. C Jurkat cells were transduced with ZSG1 VLPs produced in HEK 293T cells by co-transfection of ZSG1 VLP producer plasmids and Tat, DDX1-HA and Rev plasmids. Transductions were performed using ZSG1 VLP equivalent to 10 ng of CA using polybrene without spinoculation. All experiments were performed three times independently in triplicate. Bars indicate mean percent of transduced cells and error bars indicate standard deviation. A asterisk designates a P value less than 0.05 and a ns designates not significant
Finally, we used a reduced amount of ZSG1 VLP to transduce ~ 80% of Jurkat cells so that effects of TatFLAG, DDX1-HA or Rev co-expression could be determined. However, co-expression of Tat-FLAG, Rev or DDX1-HA had no significant effect on transduction by ZSG1-VLPs (Fig. 5C). Hence, the results overall indicate that coexpression of Tat and DDX1 together can improve the transducti on ability of NB-ZSG1 VLPs specifically.
-
Interestingly, although introduction of Nullbasic antagonist proteins improved transduction efficiency of NB-ZSG1 VLPs in Jurkat cells, this method did not improve transduction of NB-ZSG1 in primary CD4+ T cells (Fig. 6). The huTat2 intrabody did not improve transduction of primary CD4+ T cells by NB-ZSG1 lentiviral VLPs (Fig. 6A), and neither did overexpression of Tat-FLAG, Rev or DDX1-HA and co-overexpression of Tat-FLAG and DDX1-HA (Fig. 6B, C). However, ZSG1 VLPs transduced ~ 50% primary CD4+ T cells (Fig. 6B), and this transduction rate increased to 75% when polybrene was replaced by recombinant human fibronectin (retronectin) (Fig. 6C).

Figure 6. No improvement in transduction of primary CD4+ T cells with NB-ZSG1 VLPs. A Primary CD4+ T cells were transduced with ZSG1 or NB-ZSG1 VLPs produced in HEK 293T-mCh, HEK 293TmCh-huTat2 and parental HEK 293T cells. Transduction was performed using polybrene with spinoculation. B Primary CD4+ T cells were transduced with ZSG1 or NB-ZSG1 VLPs produced in HEK 293T cells by co-transfection of VLP producer plasmids and either Tat, DDX1, Rev or combination of Tat and DDX1. Transduction was performed using polybrene with spinoculation. C The same VLPs as in B were used to transduce primary CD4+ T cells using RetroNectin with spinoculation. All experiments were performed three times independently in triplicate. Bars indicate mean percent of transduced cells and error bars indicate standard deviation. A * designates a P value less than 0.05 and a ns designates not significant
In summary, NB-ZSG1 can block transduction by lentiviral gene therapy vectors. The block to efficient transduction can be mostly overcome by co-expression by TatFLAG and DDX1-HA in the VLP producer cell line. While this can improve transduction of Jurkat cells using NBZSG1-VLP, the modifications do not permit transduction of primary CD4+ cells. The results show that NB-ZSG1 is an extremely potent inhibitor of lentiviral VLP transduction especially in primary CD4+ T cells.
An Improvement in Transduction Efficiency with a Nullbasic Lentiviral Vector Using an Intracellular Antibody (intrabody) to Tat
Improved Transduction of Jurkat Cells by NBZSG1-VLPs Using Cellular or Viral Proteins
3-3. No Improvement in Transduction Efficiency of NB-ZSG1 VLPs in Primary CD4+ T Cells
-
Lentiviral vectors are useful for delivering genes of interest into mammalian cells. The ability of lentiviral vectors to transfer genes not only to dividing cells but also to nondividing cells and to maintain stable gene expression offers advantages over gammaretrovirus vectors. Lentivirus vectors are able to transduce quiescent cells because their preintegration complexes (PIC) are imported through the nuclear pore (Okitsu et al. 2003). Furthermore, lentiviral vectors are considered to be safer than gammaretroviral vectors because of reduced risks of oncogenesis (Schambach et al. 2013).
Nullbasic was delivered to T cells by an HIV-1 derived lentiviral pSicoR vector. However, Nullbasic delivery by this lentiviral vector was hindered by low transduction efficiency due to the inhibiting effects of Nullbasic on HIV-1. The fact that Nullbasic inhibits reverse transcription causes a hindrance in lentiviral VLP transduction of target cells such as Jurkat or primary CD4+ T cells (Apolloni et al. 2013). In an attempt to overcome this problem, we used viral and cellular proteins that had potential to act as Nullbasic antagonists during VLP production.
When NB-ZSG1 VLPs were produced in a HEK 293T cell line expressing the huTat2 intrabody, transduction efficiency in Jurkat cells was improved approximately two fold. Transactivation assays showed that huTat2 intrabody inhibited Tat-mediated transactivation, indicated by decrease of luciferase activity by ~ 25%, which was less than previously reported (Marasco et al. 1999). Nevertheless, the results suggest that huTat2 protein interferes with Nullbasic thereby increasing Nullbasic-VLP infectivity in Jurkat cells, however the exact mechanism by which huTat2 rescues Nullbasic-VLP transduction remains to be elucidated. Given that the level of rescue is low, defining a precise mechanism by which huTat2 opposes Nullbasic at the molecular level could prove challenging.
Overexpression of Tat-FLAG with or without spinoculation did compensate for Nullbasic inhibition of NB-ZSG1 VLP transduction of Jurkat cells. Tat has been shown to enhance reverse transcription of HIV-1 (Harrich et al. 1997; Apolloni et al. 2007). Both Tat and Nullbasic bind to reverse transcriptase (Apolloni et al. 2007; Lin et al. 2015). Thus, we hypothesized that overexpression of Tat-FLAG might compete for interaction with reverse transcriptase and that Tat-FLAG may partially or completely mitigate the negative effect of Nullbasic on reverse transcription. The results from this study suggest this hypothesis is possible.
We additionally hypothesized that transduction efficiency of T cell lines and primary CD4+ T cells may be limited to some extent by the availability of full length lentiviral mRNA in the packaging cell line. This is because Nullbasic inhibits Rev which is required for RRE-directed export of viral mRNA (Lin et al. 2012, 2014), and that overexpression of Tat or DDX1 reversed Nullbasic interference of Rev (Lin et al. 2014). DDX1 assists HIV-1-RevRRE complex formation by binding to RRE-containing viral mRNA (Fang et al. 2004; Robertson-Anderson et al. 2011). Here we showed that individual treatment of NBZSG1 VLP producer cells by DDX1-HA and Tat-FLAG overexpression together with spinoculation increased the transduction efficiency of Jurkat cells by NB-ZSG1 lentiviral VLPs. Hence, it is possible that the overexpression of DDX1-HA compensated for Nullbasic inhibition of RevRRE-NB-ZSG1 mRNA transport from the nucleus to cytoplasm. However, overexpression of Rev was not effective in improving NB-ZSG1 transduction efficiency. This is consistent with the findings of a previous report showing direct interaction of Nullbasic with DDX1-HA, but not with Rev, was a mechanism of Nullbasic inhibition of Rev function (Lin et al. 2014). The improvement in the transduction rate of Jurkat cells by NB-ZSG1 VLPs was more pronounced after combined overexpression of TatFLAG and DDX1-HA during VLP production, suggesting that these proteins have additive effects to counteract Nullbasic inhibition of both transcription and viral mRNA nucleocytoplasmic transport. When the same treatments were applied to ZSG1 VLPs, there was no significant difference in the transduction efficiency of Jurkat cells. This indicates that the effect of huTat2 intrabody, Tat-FLAG and DDX1-HA on NB-ZSG1 lentiviral VLP transduction efficiency was specific to the effects of Nullbasic on lentiviral VLP production and transduction of cells by these VLPs.
Spinoculation increased the transduction efficiency of all the tested VLPs in Jurkat cells. Spinoculation has a concentrating effect on VLPs. Also, the centrifugal pressure on the cells stimulates actin and cofilin dynamic activity (Guo et al. 2011). Actin is a protein located in the cell cytoplasm that forms microfilaments as a primary component in the cytoskeleton, while cofilin is an actinbinding protein responsible for enhancing the treadmilling of actin filaments. Spinoculation modulates actin and cofilin dynamic activity (Guo et al. 2011). VSV-G enveloped virus fusion to target cells is increased by actin filament formation. Furthermore, the actin cytoskeleton helps intracellular lentivirus trafficking (Taylor et al. 2011), and thus spinoculation improved lentivirus transduction efficiency. In some respect, the results suggest that spinoculation enabled improved transduction when DDX1-HA was overexpressed in the VLP producer cell but the reason for this is unknown.
Although the addition of hu-Tat2 intrabody, Tat-FLAG, DDX1-HA and Rev proteins as well as spinoculation improved NB-ZSG1 transduction efficiency of Jurkat cells, these methods did not help transduction of primary CD4+ T cells with NB-ZSG1 lentiviral VLPs. In contrast, up to ~ 75% transduction efficiency was achieved in primary CD4+ T cells transduced with ZSG1 VLPs. This indicates that lentiviral VLPs can transduce primary CD4+ T cells, but not when Nullbasic is present. These results indicate that Nullbasic inhibition of lentivirus was more prominent in primary CD4+ T cells than in Jurkat cells. There are many possible causes for this result, such as cell factor differences between Jurkat and primary CD4+ T-cells, which may have different levels of important cell factors required for lentiviral transduction.
Like Nullbasic, the cellular restriction factor SAMHD1 inhibits retroviral reverse transcription however by different mechanisms. For example, SAMHD1 is reported to bind DNA and RNA and to have nuclease activity (Ahn 2016). SAMHD1 inhibits HIV-1 in cells where SAMHD1 is expressed at high levels. SAMHD1 dNTPase sharply reduces cytosolic levels of dNTPs so that reverse transcription is inefficient (Ahn 2016). These activities are unlikely to be a factor here because only activated and proliferating CD4+ T cells were used in these experiments. Under these conditions SAMHD1 is reported to be down regulated (Ruffin et al. 2015).
Overall, transduction efficiency of Jurkat cells by Nullbasic lentiviral VLPs can be improved by the presence of huTat2, Tat-FLAG and DDX1-HA together with spinoculation. Therefore, these methods can be used to further study Nullbasic activities and interactions using Jurkat cells. Although lentiviral vectors are a preferred delivery system in gene therapy due to their ability to transduce non-dividing cells, provide stable transgene expression and have less oncogenic risks, currently available HIV-1-based lentiviral vectors cannot be used to deliver Nullbasic to primary CD4+ T cells. Vectors based on HIV-2 or SIV would be worth testing as the negative effects of Nullbasic may be specific. The Nullbasic inhibition of lentivirus was pronounced in primary CD4+ T cells so that addition of Tat-FLAG, Rev and DDX1-HA did not improve Nullbasic lentiviral VLP transduction efficiency. It is possible that future experiments could pursue improved expression of Nullbasic antagonists, such as Tat or DDX1, so that an optimized system may overcome the transduction block imposed by Nullbasic, especially in primary CD4+ T cells. Overall, this study supports continued development and use of gammaretroviral vectors or non-HIV-1 based lentiviral vectors, not inhibited by Nullbasic, in order to deliver Nullbasic to human primary cells.
-
This research was supported by the National Health and Medical Research Council Project Grant (1085359). LR was supported by Prime Minister's Australia Asia Endeavour Postgraduate (Ph.D.) Award funded by the Australian Government, Department of Education and Training, UQ international scholarship (UQI) and UQ Centenial scholarship (UQCent). We thank the QIMR Berghofer Flow Cytometry and Imaging Facility for technical expertise with cell sorting and flow cytometry. We thank Wayne Marasco for providing the sFvhutat2 expression plasmid for this study.
-
DH and LR originated and guided the study. DL, DH, LR and HJ designed the experiments. LR, ML and HJ performed experiments. LR, DH, HJ and DL wrote the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
"Buffy coat" human blood cells were obtained from the Australian Red Cross. All samples were provided by donors with informed consent.














 DownLoad:
DownLoad: