-
Dear Editor,
In recent years, the incidence of human infections caused by emerging or re-emerging pathogens has rapidly increased. Diseases that were once regional now have the ability to spread globally in a short amount of time and pose a wider threat to public health (Weaver et al. 2018). Yellow fever virus (YFV, family Flaviviridae, genus Flavivirus) is a mosquito-borne flavivirus that causes yellow fever in humans and has been endemic in Africa and Latin America for many years (Domingo et al. 2018). The most recent large-scale outbreak of YFV occurred in Brazil in which the mortality rate as of February 28, 2018 is 32.78% (WHO 2018). Rift Valley fever virus (RVFV, family Bunyaviridae, genus Phlebovirus) is another mosquitoborne virus and primarily circulates in Africa and the Middle East, and in recent years in Europe (Mansfield et al. 2015). During the initial stage of infection, most patients infected with YFV or RVFV present nonspecific symptoms such as fever, headache, and vomiting, which often lead to a misdiagnosis (Mansfield et al. 2015; Domingo et al. 2018). The cases of YFV and RVFV in China were first reported in March and July 2016, respectively, in travelers returning from Angola (Chen et al. 2016; Liu et al. 2017).
Humans infected with RVFV or YFV can develop viremia during the acute phase of the disease. Currently, laboratory diagnosis of YFV (Domingo et al. 2018) and RVFV (Mansfield et al. 2015) infections are mainly performed using sera tested by quantitative reverse transcription polymerase chain reaction (qRT-PCR) or immunological methods such as enzyme-linked immunosorbent assay (ELISA). YFV (Domingo et al. 2018) and RVFV (Mansfield et al. 2015) can be detected in human kidneys or kidney fibroblasts and cause pathological changes in the kidneys of patients with severe disease. Recent studies have shown that YFV may be detected in urine samples and persists in the blood (Reusken et al. 2017; Barbosa et al. 2018; Domingo et al. 2018) and that viable YFV can be isolated from urine samples (Barbosa et al. 2018). In contrast, RVFV has been detected in urine samples using qRT-PCR (Haneche et al. 2016), but the isolation of live virus has not been reported.
Here, we report results from a comparative analysis of the detection window for YFV and RVFV in patients' sera and urine samples. Moreover, infectious YFV and RVFV were isolated from clinical samples, verified by sequencing, and whole-genome sequences were compared. Between March 8 and April 10, 2016, a total of 11 YFV cases were imported into China (Wang et al. 2016). On July 21, 2016, the first imported case of RVFV was reported in China (Liu et al. 2017). Serum and urine samples were collected at different time points after disease onset from five of the YFV patients infected with a West Africa Ⅱ genotype of YFV (patients YF-BJ1, YF-SH1, YF-BJ3, YF-BJ4, and YF-BJ5) (Ling et al. 2016; Chen et al. 2018) and the RVFV patient infected with an E lineage RVFV (patient RF-1) (Supplementary Fig. S1, Supplementary Table S1). Of the five YFV-infected patients, only YF-BJ5 was vaccinated with the attenuated YFV vaccine with the vaccination occurring in Namibia 10 months prior to disease onset (Song et al. 2018) (Supplementary Table S1). Except for the death of YF-BJ1, the only fatality, the remaining patients with severe (YF-BJ3 and RF-1) or mild symptoms (YF-SH1, YF-BJ4, and YF-BJ5) were all recovered. The detection window for YFV in serum and urine was analyzed first by qRT-PCR (Supplementary Materials and Methods, Fig. S1A–S1D; Supplementary Table S1). In patient YF-BJ1, YFV was detected at comparable levels in all samples (Fig. 1A; Supplementary Table S1). For patient YF-BJ3, serum samples were positive until 10 days after disease onset (dao), whereas YFV in urine persisted until 32 dao (Fig. 1B; Supplementary Table S1), which was longer than other reports on natural YFV infections (Reusken et al. 2017; Barbosa et al. 2018). In addition, following vaccination with the attenuated vaccine strain YFV-17D, RNA was detected up to 198 days in urine post vaccination (Martins et al. 2013). In patients YF-BJ4 and YF-BJ5, the detection windows were also longer for urine compared to that for sera (Fig. 1C, 1D; Supplementary Table S1). For patient RF-1 (Liu et al. 2017), RVFV was detected in both sera and urine (Supplementary Table S1). As indicated above, our results were consistent with previous findings (Haneche et al. 2016; Reusken et al. 2017), suggesting that urine may be used as an appropriate clinical specimen for the detection of YFV and RVFV and may prove useful for instances when blood samples are not available.
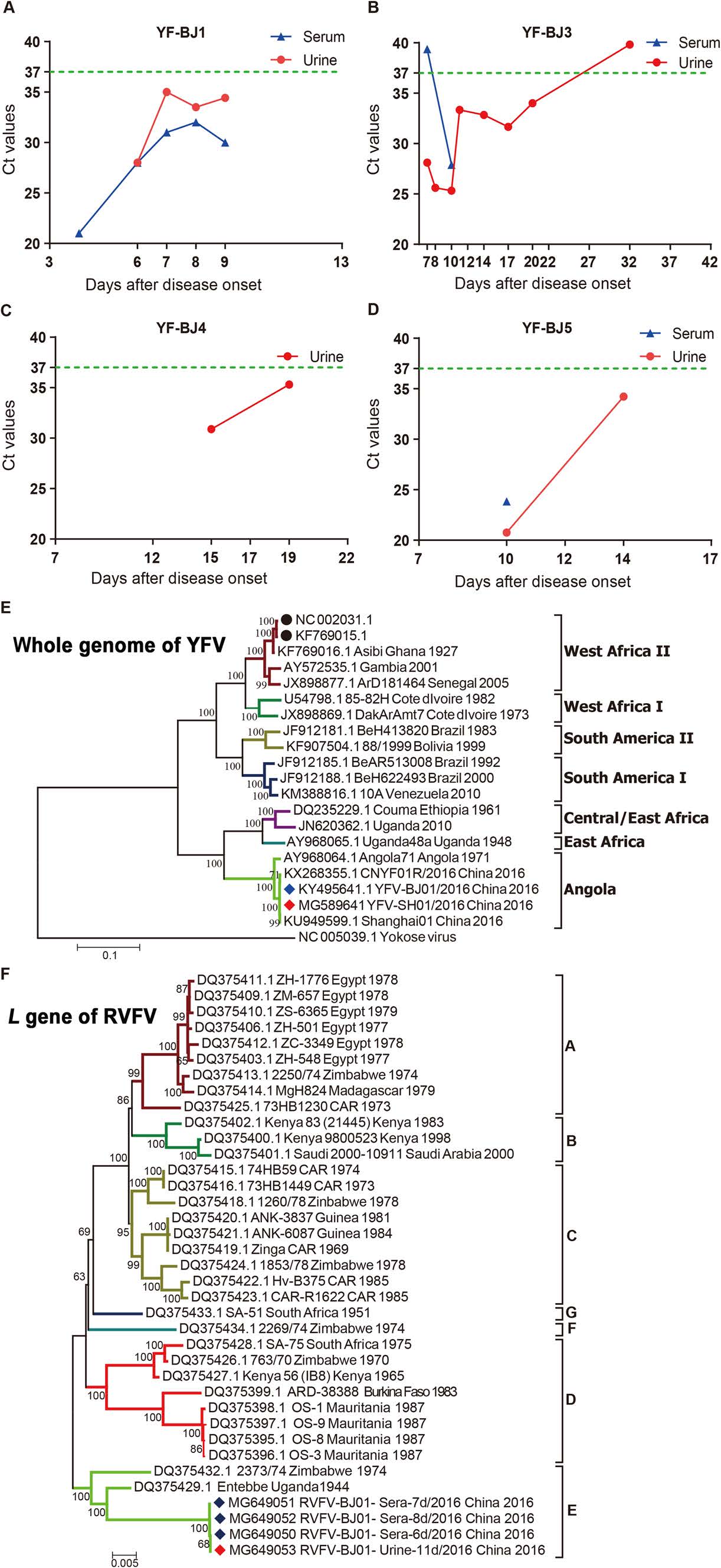
Figure 1. A–D Detection of yellow fever virus (YFV) in serum and urine specimens from YFV-infected patients (YF-BJ1, YF-BJ3, YF-BJ4, and YF-BJ5) by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The results from sera and urine are indicated in blue and red, respectively. The green dotted line represents the detection threshold by qRT-PCR. The Ct values are shown in Supplementary Table S1. E–F Phylogeny of Rift Valley fever virus (RVFV) and YFV isolates from patient sera and urine. Nucleotide sequences for the whole genome of YFV (E) and L gene of RVFV (F) were inferred by the Maximum Likelihood method with 1000 replicates using Molecular Evolutionary Genetics Analysis version 6.0 (MEGA 6). Strains indicated with a red or blue diamond represent sequences obtained from urine and serum samples, respectively, from the current study. Strains indicated with solid black circles represent YFV vaccine 17D strains. Each branch in the phylogenetic tree was named as follows: GenBank accession number, strain name, country of isolation, and year of isolation.
We then attempted to isolate infectious virus from the urine and sera of patients YF-BJ1, YF-SH1, YF-BJ3, and RF-1. Whole-genome sequences of the isolates were obtained by next generation sequencing (NGS) and used for strain confirmation (Supplementary Materials and Methods; Supplementary Table S1). For patient YF-BJ1, viable virus was successfully recovered only from serum collected on 4 dao and the isolate was sequenced (YFV-BJ01/2016, GenBank No. KY495641). YFV-BJ01/2016 shared 99.5% nucleotide identity to the full-genome sequence of strain CNYF01/2016 (GenBank No. KX268355), which was previously isolated from the sera of the same patient at 3 dao (Chen et al. 2016). For patient YF-SH1, virus was isolated from the urine sample collected on 13 dao (YFV-SH01/2016, GenBank No. MG589641) and the isolate shared 99.9% nucleotide identity to strain Chinese Shanghai01 (GenBank No. KU949599), which was previously isolated from the sera of the same patient (Ling et al. 2016). Phylogenetic analysis of the whole-genome sequences showed that both strains YFV-BJ01/2016 and YFV-SH01/2016 belonged to the Angola genotype, while vaccine strains 17D (GenBank No. KF769015 and GenBank No. NC_002031) belonged to the West Africa Ⅱ genotype (Fig. 1E). Furthermore, strains YFV-BJ01/2016 and YFV-SH01/2016 possessed only 74% nucleotide identity to full-genome sequences of 17D strains (KF769015 and NC_002031), indicating a long evolutionary distance between the prevalent circulating viruses and the vaccine strain. For patient YF-BJ3, infectious YFV was isolated in urine collected on 14 dao, but no viable virus was obtained from sera after 10 dao (Supplementary Table S1).
Viable RVFV was successfully isolated from the urine sample collected on 11 dao (GenBank Nos. MG649053, MG649057, and MG649061), consistent with the positive results from the serum samples collected at 6, 7, and 8 dao (GenBank Nos. MG649050-52, MG649054-56, and MG649058-60). Nucleotide identity was 99.8%–100% for the S, M, and L gene segments among four strains isolated from sera and urine (Supplementary Table S2). The genomic heterogeneity of YFV or RVFV from the same patient may have been due to intra-host evolution, as described previously (Chen et al. 2018). The S, M, and L gene segments of the isolates from sera clustered closely with the virus from urine and fell into Group E in the phylogenetic tree. The L gene results are shown in Fig. 1F. The sequence and assembly data of strains from patients YF-BJ1, YF-SH1, and RF-1 are available from the China National GeneBank (CNGB) Nucleotide Sequence Archive (CNSA; https://db.cngb.org/cnsa/; accession number CNP0000083). In the current study, YFV and RVFV were detected in and isolated from the urine of infected patients. In addition, several arboviruses, such as West Nile virus (WNV), dengue virus, and Zika virus (ZIKV), have been previously isolated from the urine of patients (Barzon et al. 2013; Andries et al. 2015; Zhang et al. 2016; Barbosa et al. 2018). Therefore, if urine specimens from YFV-infected, RVFV-infected, or other arboviruses-infected patients are not properly treated, there might be a potential for transmission by direct contact or by vectors. The detection of viable YFV and RVFV in urine samples implied that there may have been active viral replication occurring in the kidneys of patients resulting in potential renal pathogenicity caused by the viruses. Moreover, accumulating evidences from studies show that urine samples might provide extended window for the molecular diagnosis of flaviviruses in patients, including WNV, dengue virus, ZIKV, and YFV (Barzon et al. 2013; Andries et al. 2015; Gourinat et al. 2015; Zhang et al. 2016; Reusken et al. 2017; Barbosa et al. 2018). Hence, urine may be considered as an alternative non-invasive source of samples for clinical detection of YFV, RVFV, and other vector-borne pathogens, which should improve diagnosis, help with surveillance efforts against the spread of these viruses, and reduce the risk of secondary infections.
HTML
-
This work is supported by grants from the National Science and Technology Major Project of China (2016ZX10004222 and 2016YFC1200800), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010102), Sanming Project of Medicine in Shenzhen (SZSM20141 2003), Shenzhen Municipal Government of China (JCYJ20160427151920801) and Beijing Municipal Science & Technology Commission (Z161100000116049), and the National Natural Science Foundation of China (NSFC) International Cooperation and Exchange Program (816110193). Y.B. is supported by the NSFC Outstanding Young Scholars (31822055) and Youth Innovation Promotion Association of Chinese Academy of Sciences (CAS) (2017122).
-
The authors declare that they have no conflict of interest.
-
Informed consent was obtained from all patients for the collection and use of all clinical specimens. This article does not contain any studies with animal subjects performed by any of the authors.
Conflict of interest
Animal and Human Rights Statement
-
The clinical manifestations for the five yellow fever virus (YFV)-infected patients YF-BJ1, YF-BJ3, YF-BJ4, YF-BJ5 (Chen C. et al., 2018), and YF-SH1 (Ling et al., 2016) and the one Rift Valley fever virus (RVFV)-infected patient RF-1 (Liu et al., 2017) included in the current study have been previously reported. Timelines of quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis and virus isolations for the five YFV-infected and one RVFV-infected patients are shown in Supplementary Fig. 1. Only patient YF-BJ5 had been previously vaccinated with the attenuated YFV vaccine, which occurred in Namibia 10 months prior to disease onset. Urine and serum samples of the patients were collected at various time points after disease onset (Supplementary Table S1).
-
Viral RNA from the clinical specimens and cultured samples were extracted using a MagaBio plus Virus RNA Purification Kit (Automatic Nucleic Acid Purification System NPA-32+, BIOER, China) as previously described (Yang et al., 2017).
-
C6/36 and Vero cells (ATCC) were grown to 90% confluence in Dulbecco's modified Eagle's media (DMEM) (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS) (GIBICO, USA) and 100 IU penicillinstreptomycin (Solarbio, China) and then washed three times with PBS buffer. Sera or urine samples from the YFV-infected or RVFV-infected patients were then inoculated onto the Vero or C6/36 cells. Supernatants were collected every 3 days after inoculation for detection of virus by qRT-PCR. After sampling at each time point, an equal volume of fresh supplemented DMEM was added to the cultures.
-
The clinical or culture samples from the YFV-infected or RVFV-infected patients were analyzed by qRT-PCR using Mabsky Bio-tech Co., Ltd. (China) the detection kits and an ABI QuantStudio 7 Real-Time cycler (Applied Biosystems, Foster City, USA) according to the manufacturer's instructions. The sample was considered negative if the Ct value was greater than 37 or undetectable.
-
YFV and RVFV viral RNA extracted from cell culture supernatants were fragmented using RNase H. The RNA fragments were then processed by end repairing, A-tailing, adapter ligation, DNA size-selection, circularization, and DNA nanoball formation according to in-house standard procedures for BGI library construction for BGI Seq500 (BGI-Shenzhen, China). DNA libraries with 300-bp inserts were sequenced using single-end 100-bp mode (SE100) on an BGI Seq500 Sequencer. Approximately 3 Gb was sequenced for each sample to obtain the wholegenome sequences of the viruses. Viral genome assemblies were done using an in-house script pipeline based on YFV Angola71 strain (GenBank No. AY968064.1) or RVFV Kakamas strain isolated from sheep in South Africa in 2009 (GenBank Nos. JQ068144, JQ068143, and JQ068142). Multiple sequence alignments were done using MUSCLE (Edgar, 2004). Phylogenetic analyses were done using the Maximum Likelihood method with 1000 replicates in Molecular Evolutionary Genetics Analysis version 6.0 (MEGA 6) (Tamura et al., 2013). Nucleotide sequence similarity among the isolates was analyzed using BioEdit version 7.0.4 software (Hall, 1999).
Patients and data collection
RNA extraction
Virus isolation
qRT-PCR
Next generation sequencing and phylogenetic analysis
-
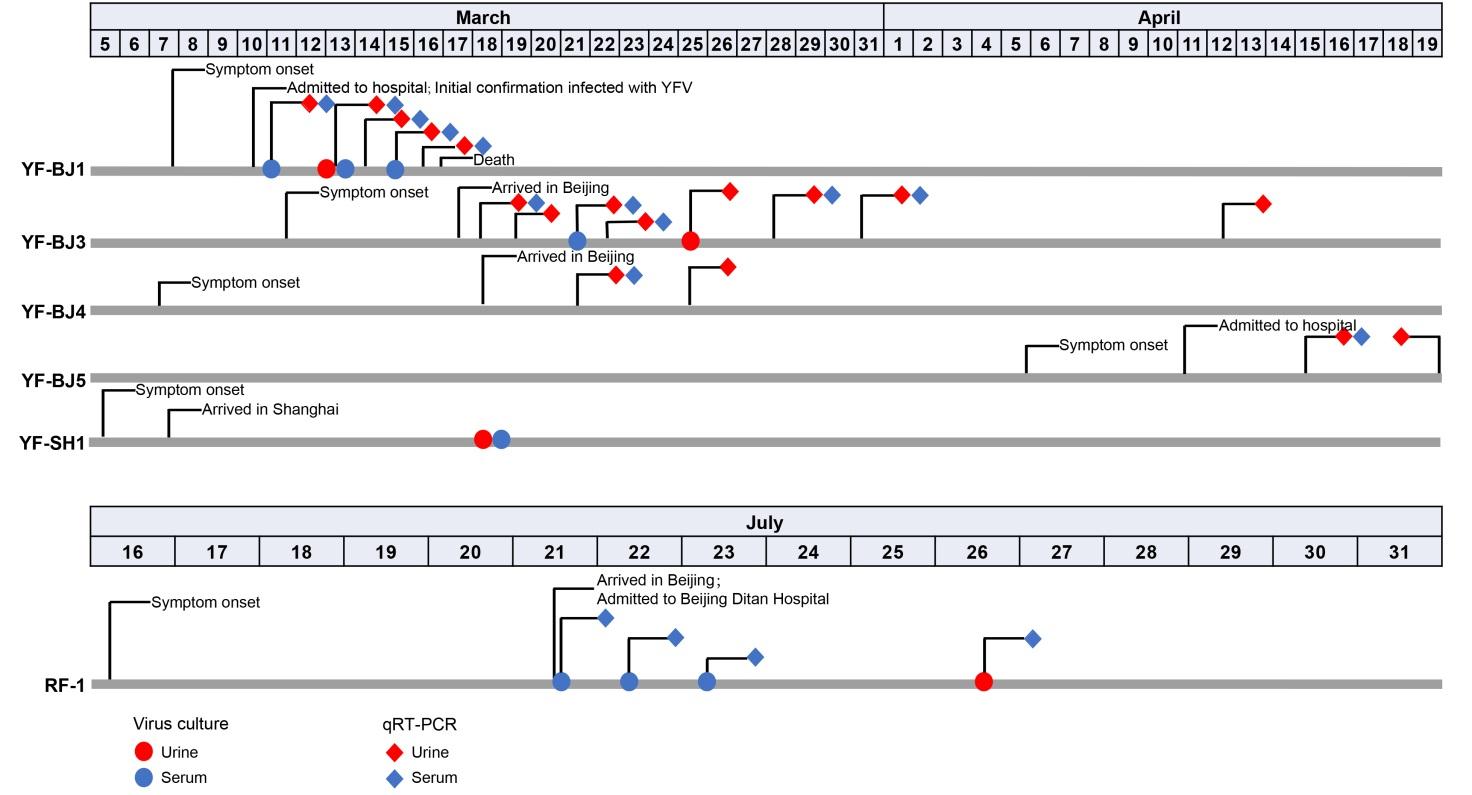
Figure S1. Clinical timelines of yellow fever virus (YFV)-infected and Rift Valley fever virus (RVFV)-infected patients. Solid red and blue circles indicate the urine and serum samples, respectively, selected for infectious virus culture. Red and blue blocks indicate the urine and serum samples, respectively, used for virus detection with qRT-PCR analysis. For patient RF-1, the Ct values for detection of virus at the different time points have been previously reported (Liu et al., 2017).

Table S1. Clinical information and viral load of the yellow fever virus (YFV)-infected and Rift Valley fever virus (RVFV)-infected patients

Table S2. Genetic identity of each gene for the RVFV isolates
















 DownLoad:
DownLoad: