HTML
-
HCV is a positive-sense, single-stranded RNA virus that belongs to the genus Hepacivirus of the family Flaviviridae. Its genome is approximately 9.6 kb in length and encodes a polyprotein that is cleaved by host and viral proteases to produce ten mature viral proteins, including structural core protein, envelope proteins E1 and E2, the p7 ion channel protein, and nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B (Yin et al. 2018). HCV infection causes acute and chronic hepatitis in humans with a high propensity for chronicity. A large number of HCV-infected patients fail to clear this virus and remain asymptomatic for years to develop severe liver diseases such as cirrhosis and hepatocellular carcinoma (Paboriboune et al. 2018). The capability of establishing chronic infection by HCV is partly due to its ability to evade host innate immune responses (Horner and Gale 2013).
RNAi is an evolutionarily conserved mechanism in all eukaryotes for post-transcriptional gene silencing. RNAi serves as an important antiviral immune response in a wide range of organisms, including fungi, plants, and invertebrates (Guo et al. 2019; Maillard et al. 2019). Moreover, recent studies have demonstrated that RNAi also exerts antiviral effects in mammals (Li et al. 2013; Maillard et al. 2013; Li et al. 2016; Qiu et al. 2017; Xu et al. 2019). The antiviral RNAi is triggered by the viral replicative intermediate double-stranded RNAs (vRI-dsRNAs) produced in the course of viral replication. These vRI-dsRNAs can be recognized and cleaved by host endoribonuclease Dicer into virus-derived small interfering RNAs (vsiRNAs) of approximately 21–23 nucleotide (nt). Argonaute (AGO) protein within RNA-induced silencing complexes (RISCs) incorporates one of the strands of vsiRNA duplex, which directs the RISC-mediated degradation of cognate viral RNAs (Maillard et al. 2019).
To counteract the antiviral RNAi, viruses have evolved to encode viral suppressors of RNAi (VSRs) targeting various steps of the RNAi pathway (Wu et al. 2010). One common strategy utilized by VSRs, including Nodamura virus (NoV) B2, Ebola virus (EBOV) VP35, influenza A virus (IAV) NS1, and human enterovirus 71 (EV-A71) 3A, is to prevent viral dsRNA from cleavage by Dicer (Li et al. 2004; Sullivan and Ganem 2005; Haasnoot et al. 2007; Qiu et al. 2017). Alternatively, multiple VSR proteins have been shown to directly target the key components of the RNAi pathway, such as Dicer or AGO proteins. For example, Wuhan nodavirus (WhNV) B2 can directly bind to Drosophila Dicer-2 to inhibit vsiRNA production (Qi et al. 2011, 2012), while Cricket paralysis virus 1A has been found to directly inhibit Drosophila AGO2 (Nayak et al. 2018).
In the case of HCV, the core protein was found to suppress RNAi by interacting with Dicer (Chen et al. 2008). Moreover, HCV structural E2 protein was also identified to antagonize RNAi by targeting AGO2 (Ji et al. 2008). Consistent with the findings in HCV, a number of RNA viruses were shown to encode more than one viral protein to counteract RNAi. For instance, VP35, VP30 and VP40 of EBOV were found to suppress RNAi through different mechanisms (Haasnoot et al. 2007; Fabozzi et al. 2011). Severe acute respiratory syndrome coronavirus (SARS-CoV) encoded 7a and nucleocapsid contain RNAi suppression activities (Karjee et al. 2010; Cui et al. 2015). Besides, Tat and Nef proteins of human immunodeficiency virus-1 (HIV-1) were identified to suppress RNAi through the dsRNA-binding and AGO interaction, respectively (Bennasser et al. 2005; Aqil et al. 2013). The phenomena of viruses encoding multiple VSRs as well as VSRs targeting different steps of the RNAi pathway highlight the importance of suppressing RNAi during the viral life cycle. Although the structural core and E2 proteins of HCV were shown to suppress RNAi, it is unclear if any HCV nonstructural protein contains RNAi suppression activity.
In this study, we uncovered that HCV nonstructural NS2 protein possessed a potent in vitro VSR activity that suppressed the RNAi induced by short hairpin RNA (shRNA) and siRNA in mammalian cells. We also confirmed the conserved residue in HCV NS2 that could disrupt its activity to suppress RNAi. Overall, our findings demonstrated that HCV NS2 contains VSR activity, thereby providing insights into the life cycle of HCV.
-
For expression of HCV nonstructural proteins including NS2, NS3, NS3/4A, NS4A, NS4B, NS5A and NS5B in HEK293T cells, their ORFs were cloned into the pRK-Flag, respectively. The templates expressing HCV proteins were kindly provided by Prof. Ying Zhu (Wuhan University, China). The point mutations were introduced into the NS2 coding region by PCR-mediated mutagenesis with the appropriate primers. For expression of recombinant NS2 in sf9 cells, the ORFs of NS2 and its mutant were constructed into the vector pFastBac HTB-MBP as previously described (Yang et al. 2017). The resulting plasmids were subjected to the Bac-to-Bac baculovirus expression system to express the fusion proteins with an MBP tag at the N-terminus. The EGFP-siRNA was chemically synthesized by Guangzhou RiboBio Co., Ltd., China. All the primers and oligonucleotides used in this study are shown in Supplementary Table S1.
-
HEK293T, 293T-NoDice (kindly provided by Prof. Bryan Cullen, Durham, NC, USA) and Huh7.5 cells were maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco) containing 7% fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin at 37 ℃ in an incubator with 5% CO2. Cells were seeded in 6-well plates and grown overnight to reach 50% confluence. Before transfection, the medium was changed to DMEM containing without serum and antibiotic. Cells then were transfected with the indicated plasmids by using FuGENE HD Reagent (Roche, Basel, Switzerland) according to the manufacturer's instructions.
-
Cells were harvested in lysis buffer [50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1% NP40, 0.25% deoxycholate and a protease inhibitor cocktail (Rhoche)]. Then the lysates were subjected to 15% SDS-PAGE and Western blotting analysis. The antibodies used in this study are as follow: anti-Tubulin (Proteintech Group, 1:5000), anti-Flag (Proteintech Group, 1:5000) and anti-Myc (Proteintech Group, 1:3000).
-
Total cellular RNAs were extracted using Trizol reagent (Thermo) according to the manufacturer's instructions. For the detection of EGFP mRNA, 5 μg of total RNAs were subjected to denatured 1.5% agarose gels with 2.2 mol/L formaldehyde. The separated RNAs were transferred onto the Hybond-A nylon membrane (GE Healthcare), and fixed at 120 ℃ for 15 min. Then the membranes were hybridized with DIG-labeled probes in Hybridization Ovens at 65 ℃ overnight. Finally, the membranes were incubated with anti-DIG antibody conjugated with alkaline phosphatase and exposed to the luminescent image analyzer LAS4000 (Fuji Film). The probes for detection of EGFP and GAPDH mRNA were complementary to their ORF region of 500–720 nt and 760–1060 nt, respectively. For detection of small RNAs, 20 μg of total RNAs were subjected to 7 mol/L urea-15% PAGE and transferred to Hybond-A nylon membrane (GE Healthcare). The membrane was chemically cross-linked in 1-ethly-3-(3-dimethylaminopropyl) carbodiimide (EDC) at 60 ℃ for 30 min. The DIG-labeled RNA probes targeting EGFP siRNA and U6 were synthesized by Takara.
-
The expression and purification of MBP alone and MBP-fusion proteins were performed in sf9 cells using baculovirus expression system as previously described (Yang et al. 2017). Briefly, cells were infected with MBP-tagged HCV NS2 expressing baculoviruses for 72 h. Infected cells were resuspended, lysed via sonication and then centrifuged at 11, 000 ×g for 30 min to remove debris. The protein in the supernatant was purified using amylose affinity chromatography (New England BioLabs, Ipswich, MA) according to the manufacturer′s protocol and then concentrated using Amicon Ultra-15 filters (Millipore, Schwalbach, Germany). All purified proteins were quantified with a bicinchoninic acid (BCA) protein assay Kit (CWBIO, China) and stored at − 80 ℃ in aliquots. Proteins were separated on 10% SDS-PAGE and visualized by Coomassie blue.
-
We generated 200-nt DIG-labeled dsRNA and 22-nt siRNA via in vitro transcription using DIG RNA labeling mix (Roche). MBP-fusion NS2 or mutant proteins were reacted with DIG-labeled RNAs (0.2 μmol/L 200-nt dsRNA or 22-nt siRNA) in a binding buffer [50 mmol/L HEPES (pH 8.0), 15 mmol/L NaCl, 0.5 mmol/L MgCl2, 10% glycerol and 1 U of RNase inhibitor (Promega)] at 22 ℃ for 45 min; the total volume was 10 μL. Then the reaction mixtures were separated on 6% (for dsRNA) or 12% (for siRNA) TBE-PAGE and transferred to Hybond-A nylon membrane (GE Healthcare). The membranes were incubated for 30 min with anti-DIG antibody conjugated with alkaline phosphatase (Roche).
-
HEK293T cells were lysed in a lysis buffer containing 20 mmol/L Tris-HCl (pH 7.4), 200 mmol/L NaCl, 2.5 mmol/L MgCl2, 0.5% Triton X100, 0.5 U/μL RNase inhibitor (Promega) and a protease inhibitor cocktail (Roche). After centrifugation for 15 min at 12, 000 ×g, the supernatant was pre-cleared via incubation with protein-A/G agarose beads (Roche) at 4 ℃ for 4 h. Then the pre-cleared lysates were incubated with antibodies (anti-Flag, anti-Myc or anti-IgG as a negative control) together with protein-A/G agarose beads (Roche) at 4 ℃ for 12 h. The antibody-bound complexes were washed for five times with the same lysis buffer except that NaCl concentration was raised to 600 mmol/L. Finally, RNAs were extracted using Trizol reagent (Thermo) according to the manufacturer′s protocol.
Plasmids and RNAs
Cell Culture and Transfection
Western Blotting
Northern Blotting
Expression and Purification of Recombinant Proteins
Electrophoretic Gel Shift Assay
RNA-IP
-
To identify whether HCV has any nonstructural protein which works as a potential VSR, we screened them via the reversal-of-silencing assay in HEK293T cells (Fig. 1A). In brief, cells were co-transfected with the plasmids encoding EGFP and EGFP-specific shRNA (shEGFP), together with the vectors for divers HCV nonstructural proteins. NoV B2 (NB2), a well-characterized VSR was used as a positive control. The expression of the HCV-encoded proteins and NB2 were detected by Western blotting with anti-Flag and anti-Myc antibodies (Fig. 1B). At 48 h post transfection (hpt), the mRNA levels of EGFP were detected by Northern blotting with a digoxigenin (DIG)-labeled RNA probe targeting 500–720 nt of EGFP ORF. EGFP-specific shRNA can effectively eliminate the EGFP transcripts (Fig. 1A, lane 2). As shown in Fig. 1A, HCV NS2 protein can effectively restore the expression of RNAi-silenced EGFP (lane 4). Expectedly, expression of NB2 or genetic ablation of Dicer (NoDice) suppressed shRNA-induced RNAi in HEK293T cells (Fig. 1A, lanes 3 and 11). Importantly, HCV NS2 did not affect the transcription efficiency of EGFP in the absence of shRNA (Fig. 1C), excluding out the possibility that HCV NS2 can directly promote EGFP transcription.
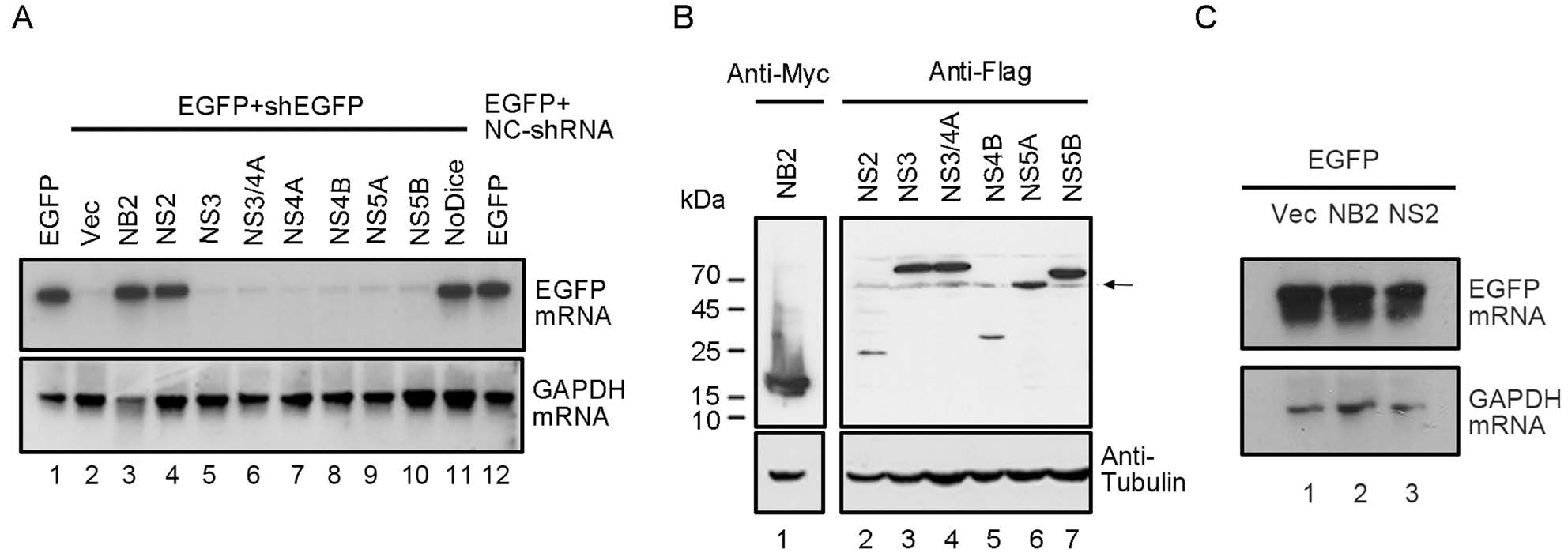
Figure 1. HCV NS2 contains RNAi suppression activity in mammalian cells. A HEK293T cells were co-transfected with the plasmids encoding EGFP (0.1 μg) and EGFP-specific shRNA (0.3 μg), together with either empty plasmid or a plasmid encoding HCV nonstructural protein as indicated or NoV B2 (NB2) (1 μg for each). 293T-NoDice cells and HEK293T cells transfected nonspecific-shRNA (NC-shRNA) were used as controls. At 48 hpt, total RNAs were extracted and the level of EGFP mRNA was examined via Northern blotting with DIG-labeled RNA probe targeting the 500–720 nt of EGFP ORF. GAPDH mRNA was used as the loading control. B The expression of HCV proteins was detected by Western blotting. Arrow indicates the unrelated bands in the bot. C HEK293T cells were co-transfected with the plasmid encoding EGFP, together with HCV NS2 or NoV B2 plasmid, respectively. At 48 hpt, EGFP mRNA levels were examined via Northern blotting.
We sought to examine whether the RNAi suppression activity of HCV NS2 is dependent on the protein expression levels. Thus, HEK293T cells were co-transfected with the plasmids for EGFP and EGFP-specific shRNA, together with the increasing amount of NS2 plasmid as indicated. Our findings showed that the reversal effect of EGFP silencing increased progressively at the mRNA levels, along with the growing amount of NS2 plasmid being transfected (Fig. 2A, 2B). We further examined the VSR activity of HCV NS2 at different time points. Our results showed that the reversal effect of EGFP silencing could be observed at 48 hpt (Fig. 2C), indicating that the VSR activity was dependent on the expression level of HCV NS2 protein. Taken together, our findings showed that HCV NS2 displays VSR activity in mammalian cells.
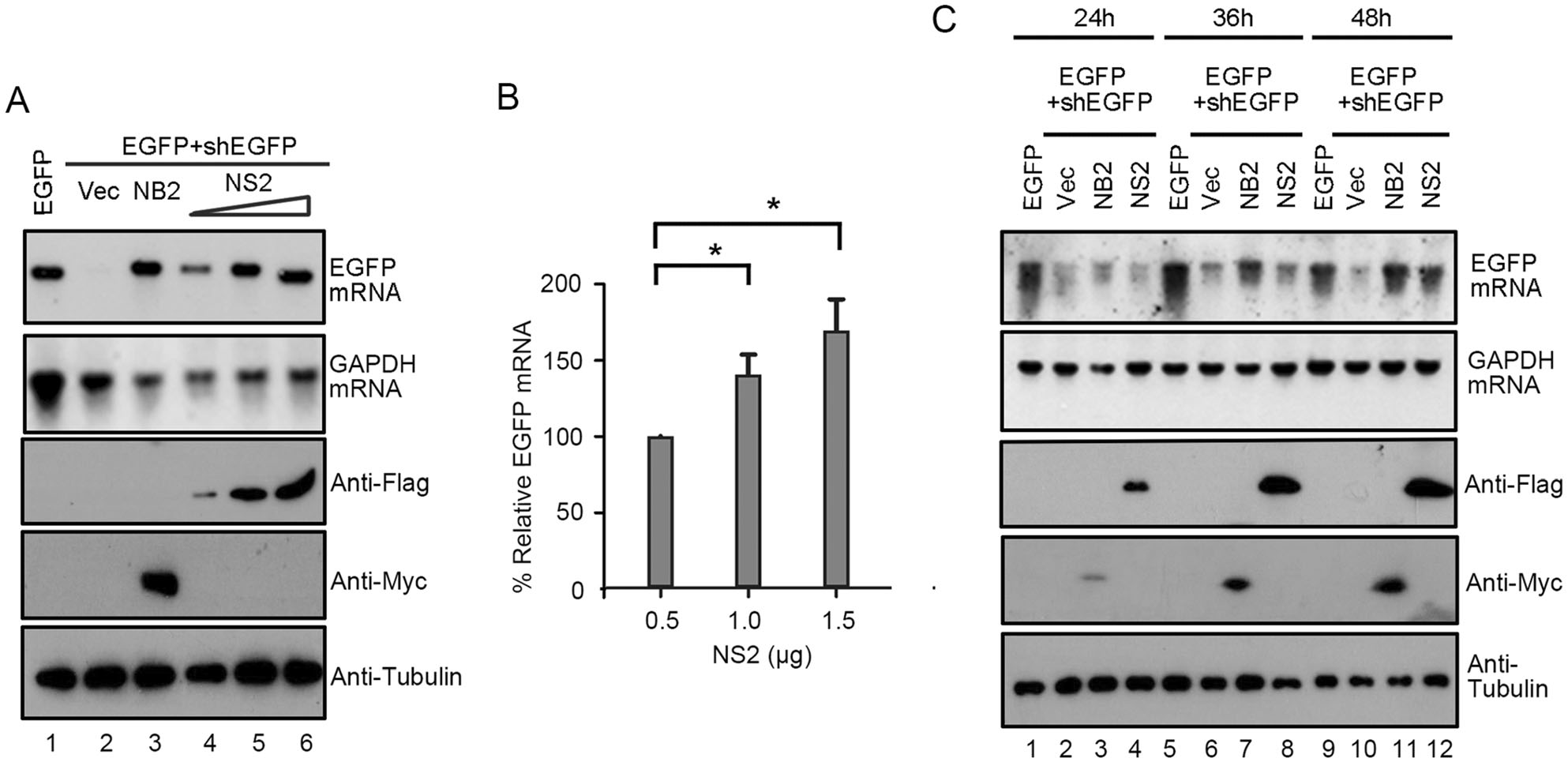
Figure 2. HCV NS2 suppresses shRNA-induced RNAi in a dose-dependent and time-dependent manner in mammalian cells. HEK293T cells were co-transfected with the plasmids encoding EGFP (0.1 μg) and EGFP-specific shRNA (0.3 μg), together with the increasing amounts of the plasmid encoding HCV NS2 (0.5 μg, 1.0 μg and 1.5 μg, respectively). At 48 hpt, EGFP mRNA levels were determined by Northern blotting (A). GAPDH mRNA was used as the loading control. The mRNA levels of EGFP normalized to that of GAPDH were quantified with Bio-Rad Quantity One software and were graphed in (B) with that in the presence of the HCV NS2 (0.5 μg) defined as 100%. Error bars represent SD values from the results of three independent experiments. *P < 0.05, as measured by two-way ANOVA (GraphPad Prism). Cell lysates were harvested and analyzed by Western blotting with anti-Flag, anti-Myc and anti-Tubulin antibodies. C HEK293T cells were co-transfected with plasmids encoding EGFP (0.1 µg) and EGFP-specific shRNA (0.3 µg), together with either empty vector or the plasmid encoding HCV NS2 (1 μg) or NB2 (1 µg) as indicated. At 24, 36, and 48 hpt, total RNAs were extracted and the level of EGFP mRNA was examined by Northern blotting. Cell lysates were harvested and analyzed by Western blotting with anti-Flag, anti-Myc and anti-Tubulin antibodies.
-
Having established that HCV NS2 contains RNAi suppression activity, we sought to examine the detailed mechanism through which HCV NS2 inhibits RNAi. In the process of RNAi, dsRNA/shRNA requires the Dicer-mediated cleavage into siRNA (Maillard et al. 2019). To investigate whether HCV NS2 can inhibit this step, small RNAs harvested from HEK293T cells co-expressing EGFP-specific shRNA together with NS2 were subjected to Northern blotting with a DIG-labeled RNA probe targeting EGFP siRNA produced from shRNA by Dicer. As shown in Fig. 3A, the accumulation of Dicer-cleaved siRNA was reduced in the presence of NS2 compared to that in cells expressing empty vector, indicating that HCV NS2 can suppress Dicer-mediated siRNA production. These results are consistent with the previous findings that HCV NS2 effectively restored shRNA-induced silencing of EGFP transcript in HEK293T cells (Fig. 1A).
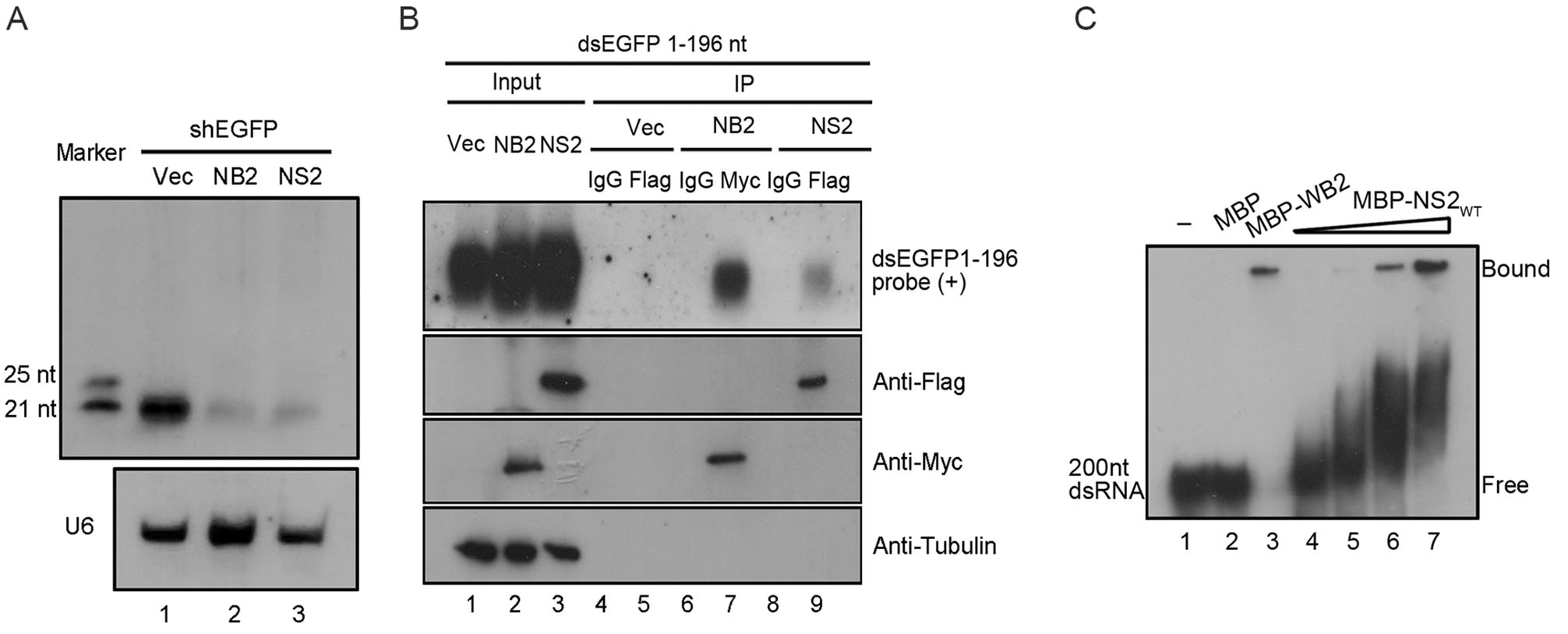
Figure 3. HCV NS2 binds to dsRNA in cells and in vitro. A HEK293T cells were co-transfected with EGFP specific shRNA (0.3 μg) and empty plasmid or a plasmid encoding HCV NS2 or NB2 (1 μg). At 48 hpt, total RNAs were extracted for small Northern blotting with either DIG-labeled oligo RNA probe targeting EGFP-shRNA or U6. The synthetic 21- and 25-nt RNAs were used as size markers. B HEK293T cells were transfected with empty plasmid or a plasmid encoding HCV NS2 or NB2 (5 μg for each). At 24 hpt, cells were transfected with the dsRNA derived from 1 to 196 nt of EGFP ORF that was transcribed by T7 RNA polymerase in vitro. Then 24 h later, cell lysates were subjected to RNA-IP with anti-Flag and anti-IgG antibody (as a negative control). Northern blotting was performed to detect precipitated NS2-bound dsRNA by an RNA probe targeting the 1–100 nt of EGFP ORF. NB2-bound dsRNA was used as a positive control (lane 7). C The increasing amount of purified MBP-NS2 protein at concentrations of 0, 2, 10, and 15 umol/L (lanes 4–7) were incubated with 0.2 μmol/L 200-nt DIG-labeled dsRNA at 22 ℃ for 45 min. Complexes were separated on 6% TBE-PAGE.
In the process of shRNA-induced RNAi, shRNA is cleaved by Dicer into siRNA. After identifying that HCV NS2 suppressed the siRNA production in cells expressing shRNA, we performed RNA immunoprecipitation (RNA-IP) assay to test whether HCV NS2 sequestrated dsRNA in HEK293T cells. Briefly, cells expressing Flag-tagged NS2, Myc-tagged NB2 or empty vector, together with EGFP-specific dsRNA (1–196 nt of EGFP ORF) were lysed and immunoprecipitated with the indicated antibodies, respectively. RNAs extracted from the RNA-IP precipitates were examined via Northern blotting with the RNA probe targeting the 196-nt dsRNA of EGFP. As shown in Fig. 3B, our findings show that NS2 can associate with dsRNA in cells (lane 9).
We sought to examine whether HCV NS2 could directly bind to dsRNA. To this end, we performed gel shift assays by incubating the recombinant MBP-fusion HCV NS2 (MBP-NS2; supplementary Fig. S1A) and the in vitro-transcribed 200-nt DIG-labeled dsRNA. As shown in Fig. 3C, the mobility of dsRNA was inhibited in a dose-dependent manner by the HCV NS2 protein (lanes 4–7), compared to the control reaction with MBP protein alone. Of note, MBP-fusion WhNV B2 (MBP-WB2), another well-characterized VSR that contains the dsRNA-binding activity, was used as the positive control (Fig. 3C, lane 3). Taken together, our findings indicate that HCV NS2 can suppress RNAi by sequestrating dsRNA.
-
We sought to examine whether HCV NS2 protein can inhibit RNAi induced by siRNA. To this end, HEK293T cells were co-transfected the plasmid for EGFP and the chemically synthesized EGFP-specific siRNA (siEGFP, 50 nmol/L), together with the NS2 plasmid. EGFP-specific siRNA induces a significant reduction of EGFP expression (Fig. 4A, lane 2). Our results showed that HCV NS2 efficiently restored the EGFP mRNA levels, indicating that HCV NS2 can inhibit siRNA-induced RNAi in mammalian cells (Fig. 4A, lane 4).
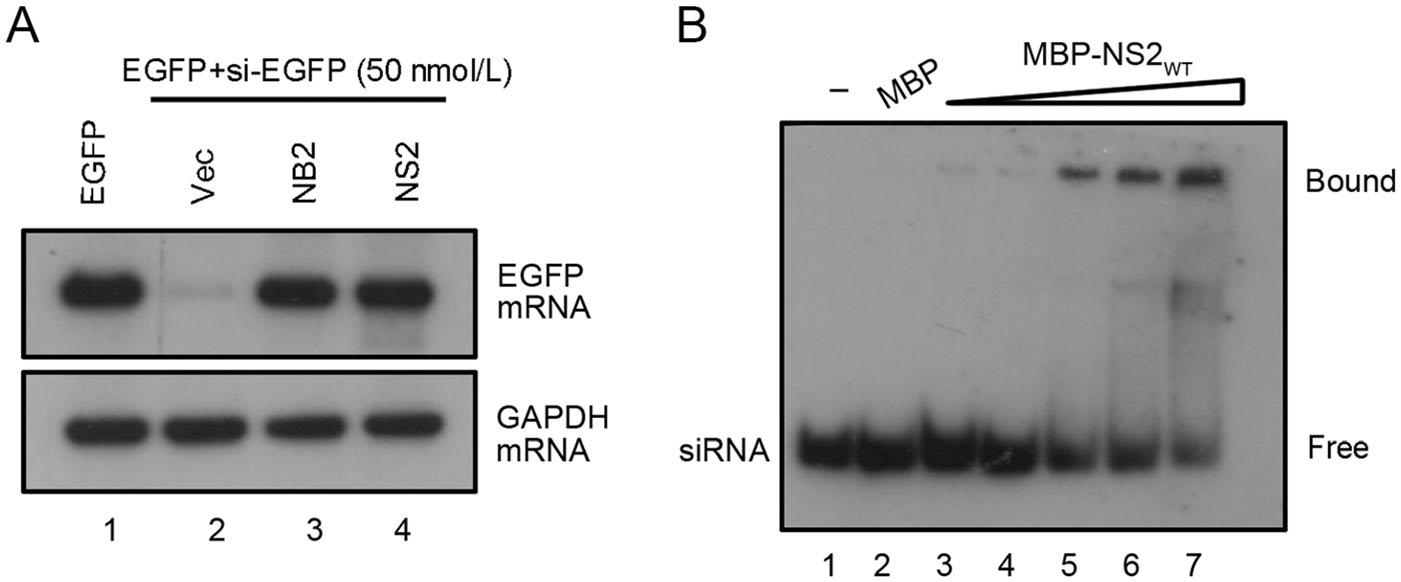
Figure 4. HCV NS2 suppresses the siRNA-induced RNAi in mammalian cells. A HEK293T cells were co-transfected with a plasmid encoding EGFP (0.1 μg) and chemically synthesized EGFP-specific siRNA (50 nmol/L), together with either empty plasmid or a plasmid encoding HCV NS2 or NB2 (1 μg for each). At 48 hpt, total RNAs were extracted for Northern blotting. GAPDH mRNA was used as the loading control. B The increasing amount of purified MBP-NS2 protein at concentrations of 0, 3, 7, 11, and 15 umol/L (lanes 3–7) were incubated with 0.2 μmol/L 22-nt DIG-labeled siRNA at 22 ℃ for 45 min. Complexes were separated on 12% TBE-PAGE.
Because HCV NS2 can directly bind to dsRNA, we sought to determine whether NS2 suppressed siRNA-induced RNAi by directly binding to siRNA. To this end, we conducted gel shift assay by incubating MBP-NS2 together with DIG-labeled synthetic 22-nt siRNA. As shown in Fig. 4B, NS2 can directly bind to siRNA in a dose-dependent manner. Taken together, these results indicate that HCV NS2 can suppress RNAi by sequestering siRNA.
-
After determining the VSR activity of HCV NS2, we sought to identify the critical residues required for its RNAi suppression activity. Thus, we performed multiple sequence alignments of NS2s encoded by multiple HCV isolates (supplementary Fig. S1B). Because the dsRNA/siRNA-binding and protein dimerization are critical for VSR's activity (Maillard et al. 2019), the positively-charged and highly conserved residues, including arginine (R), lysine (K), histidine (H), glutamic acid (E) and cysteine (C) were subjected to single-point mutation to alanine (A), and the resulting mutant NS2 proteins were then examined via the reversal-of-silencing assay in HEK293T cells (Fig. 5A–5C). Our findings show that the C184A mutation (NS2C184A) significantly suppressed the activity of HCV NS2 to suppress RNAi (Fig. 5C, lane 7 and Fig. 5D, lane 5). Moreover, we further confirmed that the C184 is critical for the VSR activity of HCV NS2 via the reversal-of-silencing assay in Huh7.5 cell line (Fig. 5E), which is usually used for hepatitis C research. Moreover, we found that C184A abolished the dsRNA- and siRNA-binding activities of HCV NS2 in vitro (Fig. 6A, 6B). Besides, we also found that NS2C184A failed to suppress the processing of shRNA into siRNA in HEK293T cells (Fig. 6C).
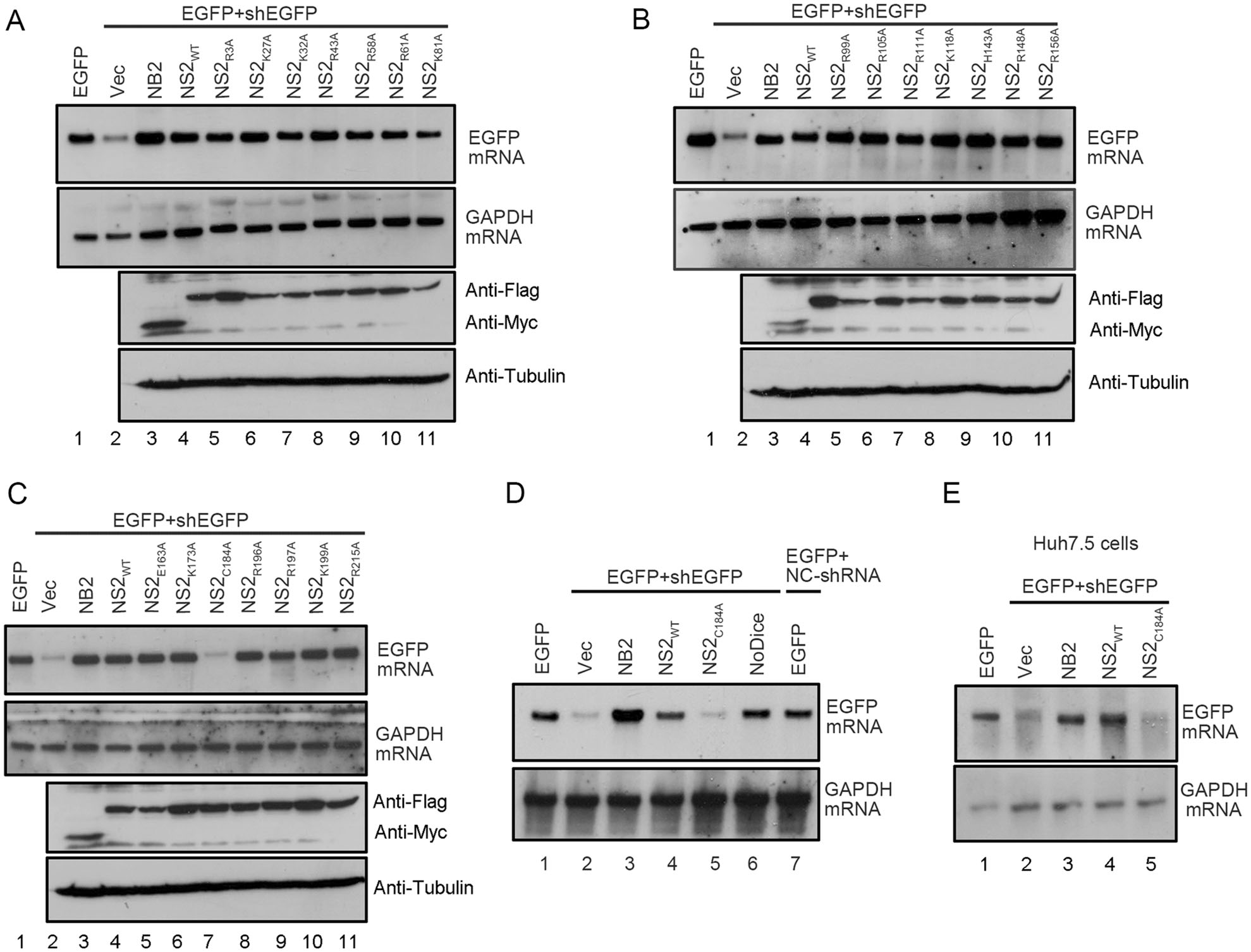
Figure 5. C184 is critical for the VSR activity of HCV NS2. A–D HEK293T cells were co-transfected with the plasmids encoding EGFP (0.1 µg) and EGFP-specific shRNA (0.3 µg), together with either empty vector or the plasmids encoding different HCV NS2 mutants (1 µg each) as indicated, At 48 hpt, total RNAs were extracted and the levels of EGFP mRNA were determined via Northern blotting. 293T-NoDice cells were used as a control. GAPDH mRNA was used as the loading control. Cell lysates were also subjected to western blotting with anti-Flag, anti-Myc and anti-Tubulin antibodies. E Huh7.5 cells were co-transfected with the plasmids encoding EGFP (0.25 µg) and EGFP-specific shRNA (0.75 µg), together with either empty vector or a plasmid encoding NS2 or NS2C184A as indicated (1.5 µg each). At 48 hpt, total RNAs were extracted and the level of EGFP mRNA was examined via Northern blotting.
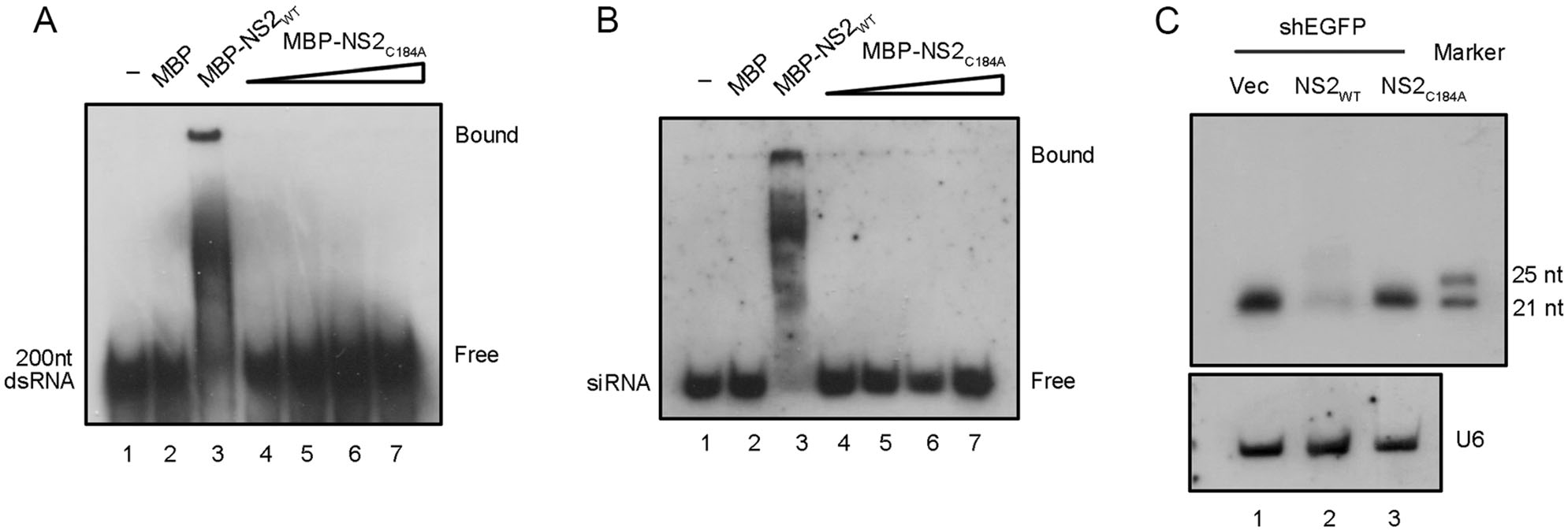
Figure 6. The dsRNA- and siRNA-binding activities of HCV NS2 are required for its RNAi suppression. MBP-NS2WT and MBP-NS2C184A were incubated with 0.2 μmol/L 200-nt DIG-labeled dsRNA (A) or 22-nt siRNA (B) at 22 ℃ for 45 min. Complexes were separated on 6% (A) or 12% (B) TBE-PAGE, respectively. C HEK293T cells were co-transfected with EGFP specific shRNA (0.3 μg) and either the plasmid encoding NS2WT or NS2C184A (1 μg of each). Total RNAs were subjected to small RNA Northern blotting. U6 was used as the loading control.
HCV NS2 has RNAi Suppression Activity in Mammalian Cells
HCV NS2 Protein Inhibits Dicer-Mediated siRNA Generation by Sequestrating dsRNA
HCV NS2 Suppresses the siRNA-Induced RNAi in Mammalian Cells
C184A of HCV NS2 is Critical for the VSR Activity
-
RNAi is an antiviral immune mechanism conserved in both invertebrates and mammals. As a counter-defense to RNAi, viruses evolved to encode VSRs that were identified to target different steps of the RNAi pathway. In the present study, we screened the viral nonstructural proteins of HCV for RNAi suppression activity and found that HCV NS2 protein is a VSR. The gel shift and RNA-IP analyses show that HCV NS2 possesses both dsRNA- and siRNA-binding activities that are required for RNAi suppression. Thus, we speculate that HCV NS2 may function as VSR by shielding virus-derived dsRNA from Dicer cleavage and binding to viral siRNA to interfere with RISC assembly.
In addition to NS2, HCV structural proteins core and E2 were also shown to contain RNAi suppression activity by directly targeting the protein components of the RNAi pathway (Chen et al. 2008; Ji et al. 2008). HCV core interacts with Dicer and inhibits the siRNA production, while HCV E2 can block the RISC assembly by interacting with AGO2. Interestingly, we showed that HCV NS2 as a VSR can target the RNA components of the RNAi pathway by directly binding to dsRNA and siRNA. These results indicate that HCV can suppress antiviral RNAi at different stages by encoding multiple VSRs. This phenomenon can also be found in other RNA viruses, such as EBOV, SARS-CoV, HIV-1 and DENV (Bennasser et al. 2005; Karjee et al. 2010; Fabozzi et al. 2011; Aqil et al. 2013; Kakumani et al. 2013; Cui et al. 2015; Kakumani et al. 2015). Thus, encoding multiple VSRs that target different steps of the RNAi pathway may offer advantages in the mode of action required for efficient suppression of RNAi, thereby highlighting the importance of the antiviral RNAi in the host-virus interactions.
HCV NS2 protein is a ~ 23 kDa transmembrane protein that has multiple functions during the HCV replication. It contains a protease domain that functions as a cysteine protease to catalyze a cleavage between NS2 and NS3 (Pieroni et al. 1997). Besides, NS2 colocalizes with E1, E2 NS3 and NS5A near the core protein and lipid droplet during the virion assembly (Ma et al. 2011). Moreover, NS2 can work as a potent interferon antagonist and apoptosis inhibitor (Erdtmann et al. 2003; Kaukinen et al. 2013). Here we found that HCV NS2 contains an RNAi suppression activity, adding a novel function to its multiple roles in the viral life cycle. HCV NS2 is identified to possess dsRNA- and siRNA-binding activities, similar to many other VSRs such as NoV B2 and SARS-CoV nucleocapsid. However, although these VSRs use the same strategy of suppressing RNAi by sequestrating dsRNA and/or siRNA, they do not share sequence identity or structural conservation, suggesting that VSRs of distinct virus families are evolved independently during the viral evolution.
HCV NS2 forms a dimer with an N-terminal alpha-helical subdomain and a C-terminal antiparallel beta-sheet (Lorenz et al. 2006). In addition, the conserved residues H143, E163 and C184 have been found to important for HCV NS2's dimerization (Lorenz et al. 2006). Our mutational analyses showed that mutation of C184 abolished the dsRNA- and siRNA-binding activities of HCV NS2 and eliminated its VSR activity, implying the dimerization of HCV NS2 is required for its RNAi suppression activity. Our findings are consistent with the previous observations that dimerization is important for VSR activities of many viruses. For example, EV-A71 3A forms a dimer and disrupting its dimerization blocks the VSR activity (Qiu et al. 2017).
In conclusion, our findings demonstrate that HCV NS2 can act as a VSR by sequestrating dsRNA from Dicer cleavage and interfering with RISC assembly by siRNA-binding. Moreover, these results add HCV NS2 as a new member of HCV-encoded VSRs, which suppressing RNAi by a different mechanism. And it provides insight into the life cycles and virus-host interactions of HCV.
-
We wish to thank Prof. Ying Zhu (Wuhan, China) and Prof. Bryan Cullen (Durham, USA) for kindly providing materials. This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB29010300 to X.Z.), the National Natural Science Foundation of China (81873964 to Y.Q., 31670161 to X.Z. and 31800140 to J.M.), the Science and Technology Bureau of Wuhan (2018060401011309 to X.Z.), and the Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory (2018ACCP-MS11 to Y.Q.). X.Z. is supported by the Newton Advanced Fellowship from the Academy of Medical Sciences, UK (NAF005\1002).
-
HZ performed the experiments, analyzed the data and drafted the manuscript; QQ, TS, JX, JK and JM helped to perform the experiments; QQ and TS helped to analyze the data; YQ and XZ designed the experiments, analyzed the data, and finalized the manuscript. All authors approved the final manuscript.
-
The authors declare that there are no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: