-
Dear Editor,
The outbreak of coronavirus disease 2019 (COVID-19) has been declared a pandemic by the World Health Organization (WHO) and has resulted in the worst public health crisis since World War II (Wang et al. 2020). The causative pathogen of COVID-19 is a novel strain of coronavirus named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2); SARS-CoV-2 is the seventh coronavirus that has been found to infect humans (Gorbalenya et al. 2020; Zhang and Holmes 2020; Zhu et al. 2020). As of June 8, 2020, more than 7 million confirmed cases of COVID-19 have been reported worldwide, resulting in approximately 400, 000 deaths (WHO 2020). As recommended by the WHO, detection and isolation are essential for containing the rapid spread of this pandemic. Therefore, adequate, large-scale screening and detection of SARS-CoV-2 are necessary. In addition, in locations in which the population has experienced and gradually recovered from COVID-19, there may be a huge demand for large-scale testing and surveillance. For example, in Wuhan city, in which the virus was successfully contained after its rapid spread, residents gradually resumed normal activities beginning in mid-March, with full re-opening of the economy by April 8, 2020. Data from Wuhan Municipal Health Commission showed that approximately 275, 400 nucleic acid tests for SARS-CoV-2 had been conducted for people who returned to work in Wuhan from April 8 to April 15 (CCTV 2020). Therefore, all countries and cities, including those still experiencing outbreaks and those in the process of recovery, are expected to require mass COVID-19 testing, which could be limited by worldwide testing capacity.
Currently, methods for detection of SARS-CoV-2 include SARS-CoV-2 nucleic acid detection, antibody (IgM/IgG) detection, and antigen detection (Corman et al. 2020; Seo et al. 2020; Xiang et al. 2020). Among these methods, nucleic acid detection is the most commonly used; the presence of viral RNAs is an indication of ongoing viral infection in the human body. At present, viral nucleic acids are generally sampled using throat swabs and detected by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). However, the capacity for SARS-CoV-2 nucleic acid detection is largely limited by the number of instruments, kits, and experienced laboratory personnel available. These limitations have caused low screening efficiency, which leads to a lag in identifying potential infections and may then exacerbate the spread of COVID-19 (Carter et al. 2020). Therefore, novel qRT-PCR approaches for nucleic acid detection are required to enhance testing efficiency.
In this study, we aimed to explore a practical method and procedure for SARS-CoV-2 qRT-PCR detection in 96-well plates using pooled throat swab samples. This method may reduce the cost of testing and increase the screening capacity for SARS-CoV-2 using existing instrument and kits, consequently facilitating the containment of COVID-19 worldwide.
The pooled sample strategy was designed owing to the low detection of infection. A similar sample pooling strategy is often used in blood donation, and a recent report on SARS-CoV-2 from the United States of America described a similar approach (Hogan et al. 2020). Because the virus from positive samples will be diluted by negative samples, the detection kit must be highly sensitive, particularly for some weakly positive samples, whose Ct values in qRT-PCR are near the critical value for distinguishing between negative and positive results. In order to identify the best SARS-CoV-2 detection kit with the highest sensitivity, we evaluated three kits (designated Kit A, Kit B, and Kit C; brand names are not included here, but can be provided upon request) that have been widely used in Wuhan. For all three kits, the ORF1ab and N genes of SARS-CoV-2 were the detection targets. The critical Ct values for Kits A, B, and C were 40, 40, and 43, respectively, and the SARS-CoV-2 detection sensitivities of Kits A, B, and C were 200, 500, and 1000 copies/mL, respectively (according to the manufacturers' instructions).
Throat swab preservation solutions from 20 COVID-19-positive patients were used in the test. Each positive sample was mixed with the throat swab preservation solution from negative subjects, and four dilution multiples (0, 5, 10, and 20) were obtained. Next, nucleic acids were extracted from each mixing tube using a nucleic acid extraction apparatus (EX2400; Shanghai ZJ bio-tech, Shanghai). qRT-PCR was then performed, and the positive detection rate for each kit was analyzed. As shown in Fig. 1A, at each dilution concentration, Kit A had the highest positive detection rate among the three kits; therefore Kit A was chosen for use in subsequent mixing saturation assays.
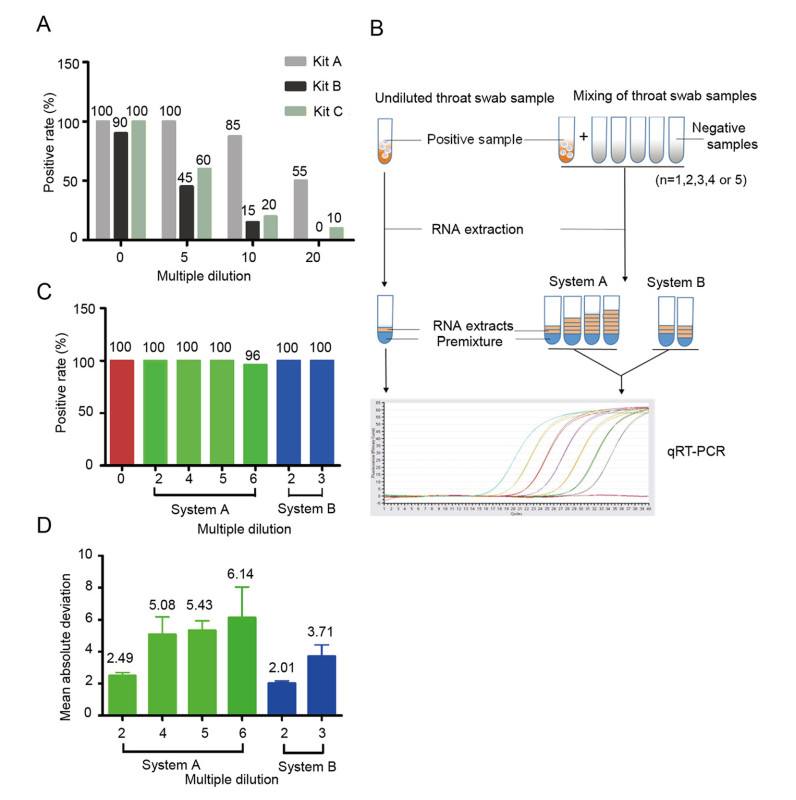
Figure 1. A Positive qRT-PCR detection rates for diluted throat swab samples using the three detection kits. Swab samples were diluted 5-, 10-, or 20-fold. B Schematic diagram of sample mixing and the two qRT-PCR systems. C Positive detection rate of diluted throat swab samples using two systems for qRT-PCR. D Mean absolute deviation of detected Ct values using systems A and B.
A new set of mixing samples was then tested, with a focus on smaller dilution multiples (2, 3, 4, 5, and 6), and 50 positive throat swab samples were evaluated. In our analysis of the Ct value distribution for the 50 original throat swabs, 8% of the sample (n = 4) ranged from 10 to 20, 48% of samples (n = 24) ranged from 20 to 30, and 44% of samples (n = 22) ranged from 30 to 40. The mixing procedure was performed as described above. To evaluate the quality and confidence of the 96-well plate qRT-PCR system, we designed and tested two reaction systems, i.e., increasing the reaction volume in system A and maintaining the system volume in system B (Fig. 1B).
In system A, the volume of the qRT-PCR premixture remained unchanged, whereas the amount of viral nucleic acid increased in the reaction system. In system B, the amount of virus nucleic acid was also increased, whereas the volume of the qRT-PCR premixture was reduced to ensure the total volume in the reaction system was maintained as 25 μL (Table 1). In both systems, dilution of throat swab samples was offset by increasing the amount of viral nucleic acid in each reaction. Our results showed that the positive detection rates for all diluted samples were greater than 96%. Notably, for samples with fivefold or less dilution, the positive detection rate was 100% (Fig. 1C). According to our analysis of the mean absolute deviation (MAD) (Geary 1936), we compared detected Ct values for the two systems with those of undiluted samples and found that the MAD values were lowest at twofold dilution in both systems A and B. As the dilution fold increased, the MAD values for the two systems also increased gradually, indicating that changes in the composition of the reaction system may decrease the reliability of the results, particularly when the reaction system is overdiluted (Fig. 1D).
Multiple dilution Undiluted
(25 µL)System A
(30–50 µL)System B
(25 µL)0 2 4 5 6 2 3 System (µL)
Mix + RNA20 + 5 20 + 10 20 + 20 20 + 25 20 + 30 15 + 10 10 + 15 Positive rate (%) 100 100 100 100 96 100 100 Mix Premixture, RNA SARS-CoV-2 nucleic acid extraction. Table 1. Two qRT-PCR systems.
In conclusion, in our study, in both systems, the amount of viral RNA in each reaction well was multiplied correspondingly after mixing the throat swab samples. Therefore, the positive samples were theoretically undiluted, which could avoid false-negative results. Our data indicated that qRT-PCR testing results were still reliable when samples were dilute to sixfold. Because the qRT-PCR test kits used in this study were not designed for testing mixed throat swab samples, this strategy can be further optimized to increase the efficiency and reliability of testing. Taken together, our results indicated that SARS-CoV-2 nucleic acid detection could be performed by mixing multiple throat swab samples, which may help lower the cost of testing and improve the efficiency of COVID-19 nucleic acid screening. Thus, this approach may have promising applications in the current COVID-19 pandemic.
HTML
-
This work was supported by the National Science and Technology Major Project (2020ZX09201-001, 2018ZX10101004 and 2018ZX10733403), and National Natural Science Foundation of China (31670161 and 31970169).
-
The authors declare that they have no conflict of interest.
-
This study conformed to the 1975 Declaration of Helsinki guidelines and was approved by the Ethics Committees of Wuhan Jinyintan Hospital (KY-2020-68.01). Written informed consents were obtained from all involved patients.














 DownLoad:
DownLoad: