HTML
-
Pseudorabies virus (PrV), the causative pathogen for Aujeszky's disease, belongs to Alphaherpesvirinae subfamily. PrV has a double-stranded DNA genome of more than 140 kb size and is packaged by the icosahedral capsid, protein tegument and lipid envelope (Engel et al. 2015). PrV can infect most mammals, including pigs, dogs, horses and rodents (Cramer et al. 2011; Kimman et al. 1991; Muller et al. 2011). Pseudorabies can be controlled using attenuated Bartha-based vaccines. However, since the latter part of 2011, pseudorabies outbreaks have occurred in many vaccinated farms in China. Phylogenetic analysis divided the Chinese strains into genotype Ⅱ (PrV-Ⅱ), and European/American strains in genotype Ⅰ, which contains Bartha-K61 (Ye et al. 2015). Notably, PrV is becoming a potential threat to the public health as recently, a human infected with this virus was reported showing symptoms of endophthalmitis. This indicates that PrV could also have a zoonotic route of transmission (Ai et al. 2018). PrV infection causes neurological disorders, respiratory disease and abortion, resulting in high fatality as well as enormous losses to the swine industry (Sun et al. 2016). The infection starts in the oropharyngeal mucosa and nasal epithelial cells (Masic et al. 1965), then, spreads to the peripheral nervous system (PNS) and eventually establishes lifelong latency via retrograde transport to the peripheral ganglia (Hafezi et al. 2012; Pomeranz et al. 2005).
Recent studies showed that non-coding RNAs play important role in virus life cycle (Damas et al. 2019). As a key member of non-coding RNAs, circRNA was first discovered in RNA viruses via electron microscopy (Wang and Fang 2018), and represented as newly identified singlestranded non-coding covalently closed circular molecules existing in almost all species (Salzman et al. 2012). CircRNAs are comparatively more stable than linear RNA by resisting RNase mediated degradation (Jeck et al. 2013; Qu et al. 2015). The diverse biological functions of circRNA were continuously discovered from the past decade (Li et al. 2018a, b). Researchers found that circRNAs could act as miRNA sponges and regulate gene expression in distinct physiological and pathophysiological conditions (Li et al. 2017a, b). Studies suggest that circRNAs might act as diagnostic or predictive biomarkers (Meng et al. 2017). Furthermore, circRNAs might also play role in cancer progression (Guarnerio et al. 2016), physiological development (Liu et al. 2019) and immune response (Chen et al. 2017; Li et al. 2017a, b). The expression pattern of circRNAs induced by simian virus 40 had been investigated previously (Shi et al. 2017), Circular RNAs were also found to be involved in antiviral signaling pathway (Ma et al. 2018; Tagawa et al. 2018). But, still the circRNAs expression profiles in PrV infected cells has not been studied in detail.
To investigate the expression profiles of host circRNAs, microRNA (miRNA) and mRNA during PrV-Ⅱ infection, high-throughput RNA sequencing was performed on the samples infected with virulent and attenuated PrV-Ⅱ strain compared with mock controls. The expression levels of 13 randomly-selected circRNAs were confirmed by RT-qPCR. The competing endogenous RNA (ceRNA) network and co-expression network was constructed. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed RNAs were also performed to predict their potential role during the PrV infection. The results of the present study will be useful for a better understanding of the mechanisms underlying the interaction between the PrV and host immune system.
-
PK-15 cells, maintained in our laboratory, was cultured at 37 ℃ and 5% CO2 in Dulbecco's modified Eagle's medium (Gibco, CA, USA) containing 10% heat-inactivated fetal bovine serum (Biological Industries, Israel) and antibiotics (100 μg/mL streptomycin and 100U/mL penicillin). The virulent PrV-Ⅱ strain DX (PrV-DX) and attenuated gE/TK gene-deleted strain gE-TK-PrV, was constructed in our laboratory (Jin et al. 2020) (US Patent No.15918547).
-
The PK-15 cells were infected with PrV-DX and gE-TK-PrV (MOI = 5) for 11 h. Total RNA was isolated from infected as well as uninfected cells using SuPerfecTRITM Total RNA Isolation reagent (Pufei, Shanghai, China) according to the manufacturer's instructions. Agarose gel electrophoresis was performed to detect the degradation of total RNA. The concentration and purity of RNA were checked by the Qubit R RNA Assay Kit (Life Technologies, CA, USA) and Nanodrop (Thermol Fisher, USA).
-
Three biological replicates of the virus infected and control samples were used for miRNA, circRNA and mRNA sequencing. Sequencing libraries of circRNA were created after the depletion of rRNA and linear RNA by NEBNext Ultra Directional RNA Library Prep Kit for Illumina according to the manufacturer's instructions. The miRNA sequencing libraries were generated by using the Small RNA Sample Pre Kit (Illumina, San Diego, CA, USA) and the mRNA were enriched by magnetic beads containing Oligo(dT). After the sequencing libraries were qualified, RNA-seq was performed using an Illumina HiSeq to get the raw data (raw reads). To get the clean data (clean reads), reads containing poly-N or adapter sequence and reads of low-quality were removed. Paired-end reads were aligned to the pig genome. CircRNA were identified using find_circ (Memczak et al. 2013) and miRNA were identified using bowtie (Langmead et al. 2009) tools, respectively. Differentially expressed (DE) transcripts were assigned significant when the P value < 0.05. The relative circRNAs and miRNAs amplification was estimated by using transcript per million tags (TPM) normalization (Zhou et al. 2010). DEseq 2 (Love et al. 2014) was used to determine the DE circRNAs and DE miRNAs with the P-value < 0.05.
-
The reliability of RNA-seq data was validated by RT-qPCR for the selected circRNAs. Briefly, RNA was isolated from virus infected cells and digested by RNaseR for 1 h according to the instruction. The reverse transcription kit was used to synthesize cDNA according to the manufacturer's instruction. The RT-qPCR was performed in a 20 μL reaction volume, including 10 μL SYBR Green Master Mix, 0.4 μL PCR Primer (forward and reverse respectively), 7.2 μL nuclease-free water and 2 μL cDNA. The reaction was carried out at 95 ℃ for 3 min, followed by 95 ℃ 10 s and 60 ℃ 34 s for 45 cycles. GAPDH was used as a reference house-keeping gene. Three independent wells were performed for each sample. Primers used in RT-qPCR are shown in Supplementary Table S1. (Thermol Fisher, USA).
-
GO (http://www.geneontology.org/) analysis was performed using GOseq to annotate the genes under the category of cellular component, biological process and molecular function (Young et al. 2010). Kyoto encyclopedia of genes and genomes pathway enrichment using hypergeometric test was also conducted to predict the involvement of cellular pathways targeted by ncRNAs expressed during PrV infection (Kanehisa et al. 2008). The pathways of GO and KEGG with the corrected P-value < 0.05 were chosen to be significantly enriched.
-
The miRanda and psRobot were used to search the potential miRNA response elements (MREs) of mRNA and circRNA. The same miRNA binding site on both mRNA and circRNA was presumed to be a potential circRNA-miRNA-mRNA interaction axis. Cytoscape was used to construct the ceRNA network.
Cells Culture and Viruses
Infection and RNA Extraction
RNA-seq and Data Analysis
Quantitative Real-Time PCR Analysis
GO and KEGG Pathway Analysis
Competing Endogenous RNA (ceRNA) Network Analysis
-
To determine whether circRNAs and miRNAs are involved during the PrV-Ⅱ infection, the PK-15 cells infected with a virulent PrV-DX strain and the attenuated gE-TK-PrV strain together with the uninfected control group were analyzed using RNA-seq. Data were processed using bioinformatics software DESeq 2 and TPM by the criteria of P-value < 0.05 considered as significant. The results showed that there were total 233 down-regulated and 216 up-regulated circRNAs in PrV-DX infected cells, 331 down-regulated and 247 up-regulated circRNAs in gE-TK-PrV infected cells and the 459 DE circRNAs (164 down-regulated and 295 up-regulated) between the PrVDX and gE-TK-PrV infected cells (Fig. 1A and 1B). The source and target genes of DE circRNAs between PrV-DX and gE-TK-PrV infected cells were intersected with the DE mRNAs (Supplementary Figure S1A). Genomic features and length distribution of novel circRNAs has been shown in Fig. 1C. Results showed that most of the circRNAs were encoded from the intron regions having size less than 2000 nt. DE miRNAs analysis of PrV-DX infected cells showed 44 down-regulated and 49 up-regulated miRNAs containing 7 and 10 novel miRNAs from each of them respectively (Supplementary Figure S1B and S1C). In case of gE-TK-PrV infected cells, there were 52 down-regulated and 43 up-regulated miRNAs with 8 and 10 novel miRNAs from each of them respectively. A total of 12 miRNAs, of which 6 were down-regulated with 4 novel and 6 were up-regulated with 1 novel miRNA, were differentially expressed in PrV-DX versus gE-TK-PrV infected cells (Supplementary Figure S1B and S1C).
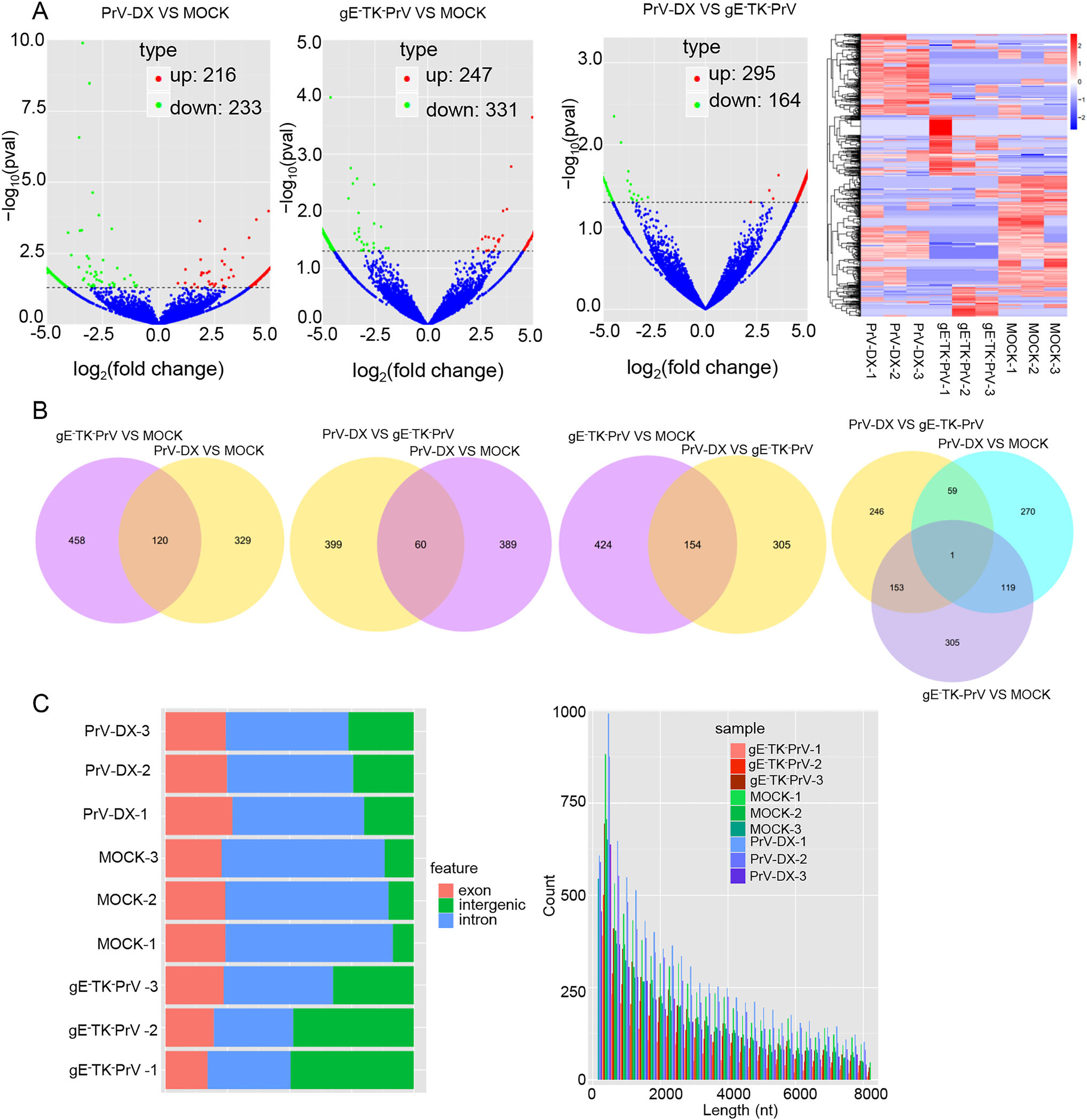
Figure 1. Differentially expressed (DE) circRNAs in PrV-DX and gE-TK-PrV infected PK-15 cells using RNA-seq. A Volcano plots of DE circRNAs. The red dots correspond to up-regulated circRNAs, the green dots correspond to down-regulated circRNAs and the blue dots represent the circRNAs without statistically significant difference. Heatmap. Clustering map shows the differential expression profile of circRNAs among different groups. B Venn diagram of the DE circRNAs in and among different groups. The intersections of the circles represent the simultaneous dyregulated transcripts in different groups. C Genomic feature and length distribution of novel circRNAs. The red part is exon, green part is intergenic, and blue part is intron.
-
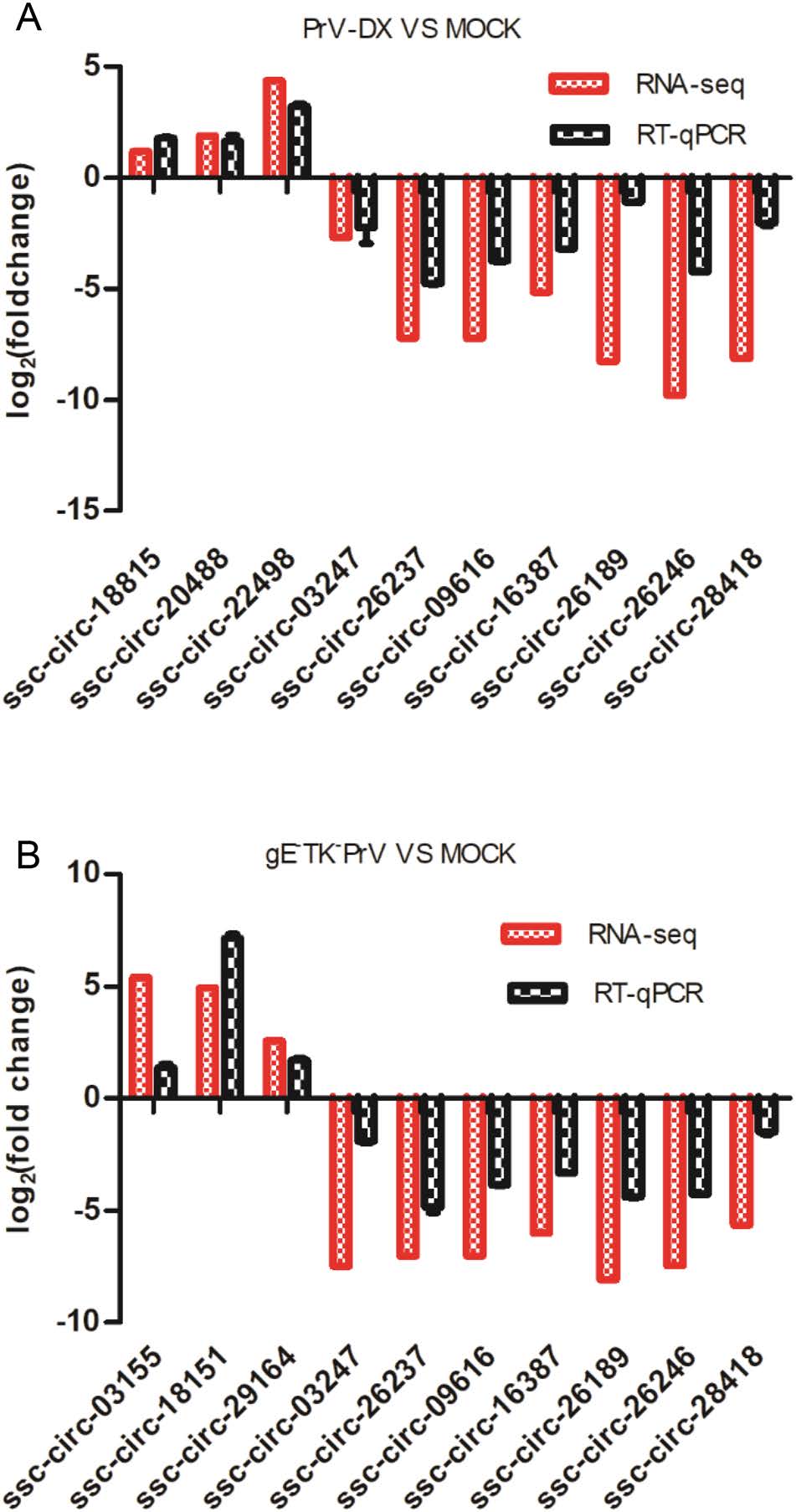
To verify the results of high-throughput sequencing, representative circRNAs expression levels were detected by RT-qPCR. The ssc-circ-18815, ssc-circ-20488, and ssccirc-22498 from PrV-DX infected cells, and ssc-circ-03155, ssc-circ-18151, and ssc-circ-29164 from gE-TK-PrV infected cells were significantly up-regulated compared to the uninfected control. Whereas, ssc-circ-03247, ssc-circ- 26237, ssc-circ-09616, ssc-circ-16387, ssc-circ-26189, ssccirc-26246, ssc-circ-28418 were down-regulated in both of the PrV-DX and gE-TK-PrV infected PK-15 cells (Fig. 2). The RT-qPCR results of the selected circRNAs were consistent with the results of RNA-seq. In addition, the expression of several selected DE miRNAs (ssc-let-7a, miR-146b, miR-210, miR-194a and miR-340 and miR-21) and mRNAs (MAP3K1 and ZNF10) were also validated and were consistent with the results of RNA-seq (Supplementary Figure S2).
-
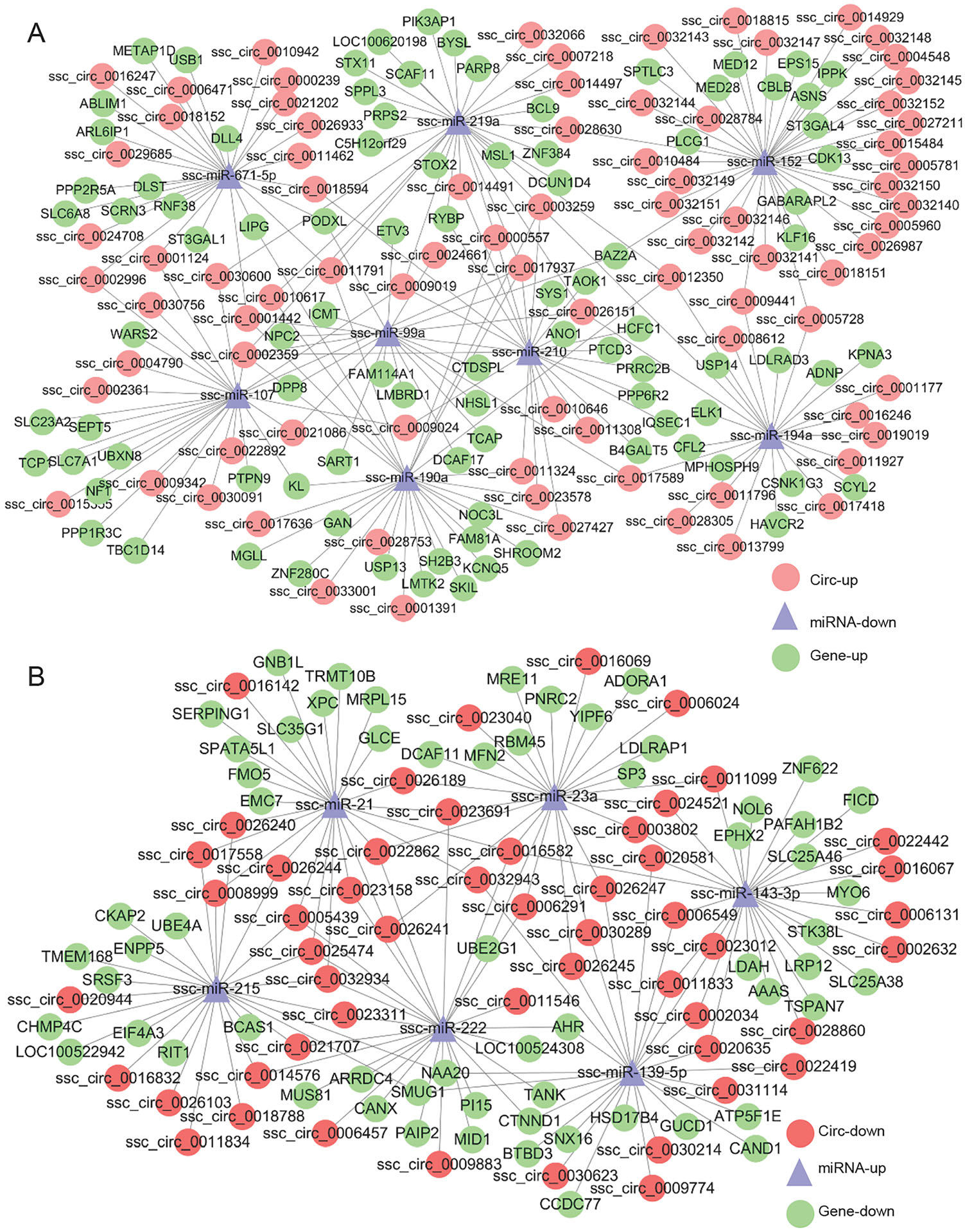
To predict the potential functions of circRNAs, a coexpression network of circRNA-miRNA-target gene was constructed using Cytoscape tool. Within the ceRNA network, circRNA indirectly regulate the expression of target genes by regulating the miRNA through their microRNA response elements (Hansen et al. 2013). Of 14 detected miRNA-mediated circRNA-mRNA competing triplets, 8 miRNAs were down-regulated (Fig. 3A) and 6 were upregulated (Fig. 3B). In addition, in the network between virulent and attenuated PrV infected cell's ncRNAs there were 5 up-regulated (Supplementary Figure S3A) and 2 down-regulated miRNAs (Supplementary Figure S3B). These miRNAs such as miR-139-5p, miR-143-3p, miR- 146b, miR-215, miR-194a, miR-210 and miR-340 (Curtale et al. 2013; Hoshina et al. 2016; Hou et al. 2018; Tian and He 2018; Wang et al. 2017a, b; Wu et al. 2012; Zhang et al. 2010; Zou et al. 2016), have possible roles in antiviral immune response.
-
GO and KEGG pathway enrichment was performed to predict the gene function of DE circRNAs and miRNAs. The target genes from the interaction network of circRNAmiRNA-target gene were used in this analysis. Three different categories, biological process (BP), cellular component (CC) and molecular function (MF) of GO annotation were taken into account (Fig. 4). The identified CC terms were intracellular, intracellular part, organelle, membrane-bounded organelle and intracellular organelle. The identified BP terms were metabolic process, cellular metabolic process, primary metabolic process and organic substance metabolic process. The identified MF mainly included protein binding and nucleic acid binding. The GO analysis of DE miRNAs was similar to those of DE circRNAs (Supplementary Fiureg S4).
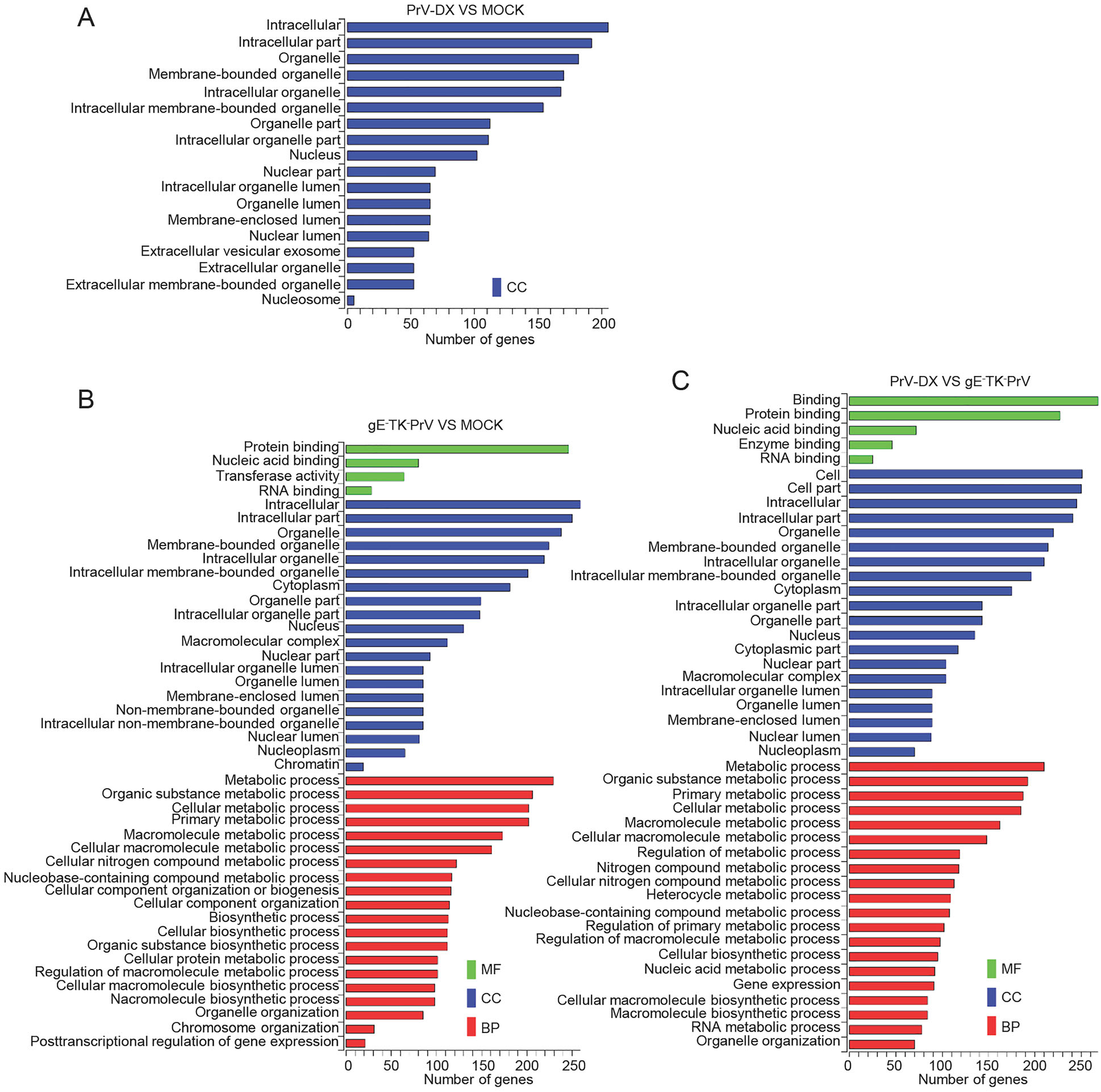
Figure 4. GO analyses of Differentially expressed circRNAs. GO term from biological process (BP), cellular component (CC) and molecular function (MF).
As a major public database on pathways, KEGG analysis was used to predict the biochemical pathways targeted by the DE circRNAs. Results showed that the DE circRNAs from infected and mock cells (Fig. 5A and 5B) involved in the pathways such as RNA degradation and regulation of actin cytoskeleton. DE circRNAs from PrV-DX and gE-TK-PrV infected cells (Fig. 5C) involved in the pathways such as Wnt signaling pathway and tight junction. The KEGG analysis of DE miRNAs were shown in Supplementary Figure S5.
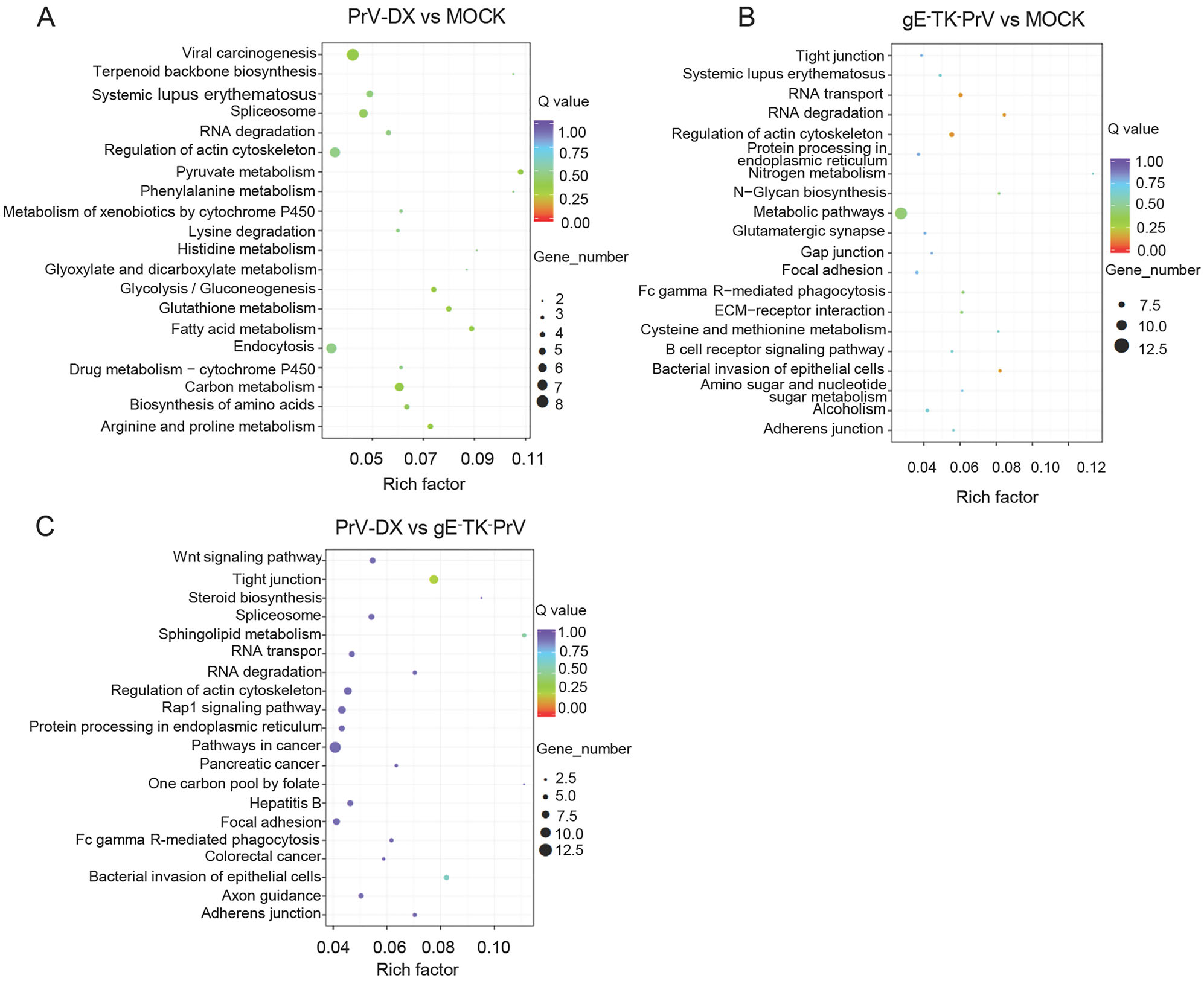
Figure 5. KEGG enrichment analyses of differentially expressed circRNAs. A KEGG enrichment analyses of DE circRNAs from PrV-DX versus MOCK. B KEGG enrichment analyses of DE circRNAs from gE-TK-PrV versus MOCK. C KEGG enrichment analyses of DE circRNAs from PrV-DX vs gE-TK-PrV.
To investigate more about the virus virulence, the intersection of the source and target genes of DE circRNAs from PrV-DX and gE-TK-PrV infected cells were enriched by GO and KEGG analysis (Supplementary Figure S6A and S6B). Results showed that intracellular, intracellular part, metabolic process were mainly enriched by GO analysis. Regarding to the result of KEGG analysis, pathways such as Wnt signaling pathway, tight junction and TGF-beta signaling pathway are enriched.
Differentially Expressed CircRNAs and miRNAs
Validation of the Differentially Expressed Transcripts by RT-qPCR
The Interaction Network of CircRNA-miRNA-Target Gene
GO and KEGG Analysis of DE CircRNAs and miRNAs
-
PrV is considered to act as a model virus to study of herpesvirus biology (Enquist 1999). Due to its possible zoonotic transmission (Yang et al. 2019), PrV-Ⅱ got attention in recent years. Previous studies indicated that the noncoding RNAs including miRNAs and lncRNAs play important role during PrV infection (Huang et al. 2014; Nishitsuji et al. 2016). CircRNAs are important member of the class of non-coding RNAs, having critical roles in regulation of gene expression (Li et al. 2015), disease progression (Peng et al. 2017) and immune response (Li et al. 2017a, b). During the infection of herpes simplex virus 1, there is a change in the expression profiles of host circRNAs (Shi et al. 2018). However, the expression profile alteration of host circRNAs and the circRNA-miRNA-mRNA axis after PrV-Ⅱ infection has not been studied so far. In the present study, 449 and 578 DE circRNAs were detected in virulent PrV-DX strain and gE and TK genes deleted attenuated strain of gE-TK-PrV infected cells respectively. Similarly, 93 and 95 miRNAs as well as 7199 and 7149 DE mRNAs were also detected. Notably, the DE transcripts between virulent and attenuated strains may be useful to understand the role of ncRNAs in determining the virulence property of PrV-Ⅱ strain. The intersection of the source and target genes of DE circRNAs from PrV-DX and gE-TK-PrV infected cells (Supplementary Figure S5A) show that most of the source genes are included in the target genes. This accord with one of the circRNA characteristics: regulating the transcription of their parental genes (Li et al. 2015). In addition, not all the source and target genes are differently expressed, indicating that except for the regulation of source and target genes, these DE circRNAs may play roles by other means. For example, they may encode a protein (Zhang et al. 2018a, b), interact with a protein (Yang et al. 2017) or change the expression of other genes (Zhang et al. 2017). To the best of our knowledge, this is the first comprehensive report about the DE circRNA, miRNA and mRNA during PrV infection.
The GO annotation analysis indicated that the DE circRNAs and miRNAs in PrV-Ⅱ infected cells involved in the protein binding, nucleic acid binding and transferase activity, which was similar to those of DE circRNAs identified during hepatitis B virus (Wang et al. 2018) and human papilloma virus (Wang et al. 2017a, b) infected cells. Actually, the function of protein binding was critical for viral infection (Sathiyamoorthy et al. 2017). These results indicated that DE ncRNAs might contribute to PrV-Ⅱ infection. Furthermore, KEGG analyses showed that the pathways of RNA transport, RNA degradation and regulation of actin cytoskeleton were specifically enriched in infected cells compared to the uninfected control cells. This suggests that the DE transcripts might be involved in the replication of viral genome.
The intersection of the source and target genes of DE circRNAs from PrV-DX and gE-TK-PrV infected cells were also enriched by GO and KEGG analysis (Supplementary Figure S5C). Metabolic process, intracellular and intracellular part are mainly enriched by GO analysis. For KEGG analysis, such as Wnt signaling pathway, Tight junction and TGF-beta signaling pathway are enriched. According to the report: Wnt signaling was demonstrated to implicate in the Virulence of Influenza Virus (Forero et al. 2015); Tight junction may influence the virus infection by affecting the entry of virus (Luo et al. 2017); TGF-beta signaling pathway can regulate INF and the function of Mitochondria (Grunwell et al. 2018). Except the displayed pathways, other pathways such as Apoptosis (Pontes et al. 2016) and NF-kappa B signaling pathway (Diel et al. 2011) were also enriched and were related to virus virulence. The results remind us that the DE circRNAs between PrV-DX and gE-TK-PrV may affect the virulence of PrV-Ⅱ by changing the epigenetic modification, signal transduction or metabolic process in these pathways.
An important function of circRNA is acting as the miRNA sponges and modulating the activity of miRNA on their target genes. In detail, miR-21, which was demonstrated to restrain PrV replication (Huang et al. 2014), was up-regulated in the virulent PrV-DX compared to attenuated gE-TK-PrV infected cells (Fig. 3), suggesting that it may relate to the viral virulence. Additionally, ssc-let-7a, which was up-regulated during PrV infection, may alter the expression of MAP3K1 and ZNF10 genes, which have role in apoptosis (Cao et al. 2015) and interact with NF-rB (Nishitsuji et al. 2015). DNM1L and SOD1, the potential target genes of ssc-miR-146b, have role in mitophagy (Zhang et al. 2018a, b) and neuropathology (Yang et al. 2014). Moreover, ssc-miR-210 (Guethlein et al. 2007), sscmiR-194a (Li et al. 2018a, b) and ssc-miR-340 (Zhou et al. 2016), which were down-regulated during viral infection, may bind to the DE circRNAs and participate in the antiviral, viral replication or immune response. Hence, the detailed information about the target genes of DE circRNAs might be useful to understand the virulence factors of PrV-Ⅱ or the process of viral infection. These results will be useful to understand the role of circRNAs during PrV pathogenesis.
To conclude, the profiles of DE noncoding RNAs and their target genes in PrV-DX and gE-TK-PrV infected PK-15 cells were analyzed by high-throughput RNA sequencing. The DE circRNAs were identified and their potential functions were predicted by GO, KEGG and circRNA-miRNA-target gene interaction network, indicating that the DE circRNAs might play critical roles during the viral infection. These results will be useful for understanding the effects and mechanisms of circRNAs in PrV pathogenesis and host response.
-
We thank Dr. Tianqi Yu for the assistance of data analysis. This work was supported by the National Key Research & Development Program of China (2016YFD0500102), Key Research & Development Program of Zhejiang Province (Grant No. 2020C02011) and the Fundamental Research Funds for the Central Universities (2017FZA6018).
-
JZ designed the study and supervised the experiment. YJ and YY prepared the samples. HL validated and analyzed the RNA-seq data and drafted the manuscript. WD and WT helped to analyze the data. JZ finalized the manuscript. All authors read and approved the final version of the manuscript.
-
The data of RNA-seq was deposited in the Gene Expression Omnibus (GEO) database with the Accession Number of GSE139424.
-
The authors declare no conflicts of interest.
-
This article does not contain any studies with human or animal subjects.
















 DownLoad:
DownLoad: