HTML
-
Although Zika virus (ZIKV) was discovered 70 years ago (Dick et al. 1952), the virus has not caused a large-scale outbreak or serious infections and has therefore received little attention. The massive ZIKV outbreak in Brazil in 2015 linked ZIKV infections with severe neurological complications such as Guillain-Barré syndrome (GBS) in adults and congenital malformations in the fetuses of women infected with ZIKV during pregnancy (Petersen et al. 2016; França et al. 2016). Because of this, ZIKV caused a public health emergency of international concern (Gulland 2016; Chitti et al. 2016) and attracted universal attention.
To date, no specific treatment or licensed vaccine for ZIKV infection is available. So specific antiviral therapies and vaccines are urgently required. Virus reverse-genetic systems are powerful tools for the development of therapies and vaccines against ZIKV. Due to the instability of the cloned flavivirus genome (Pu et al. 2011; Zheng et al. 2016; Münster et al. 2018), it is difficult to construct a plasmid containing a full-length genomic infectious flavivirus clone; however, scientists have successfully rescued recombinant ZIKV viruses using different reversegenetic systems and strategies. These include the transfection of in vitro-transcribed genomic RNA derived from plasmids containing the T7 promoter (Shan et al. 2016; Yang et al. 2017; Münster et al. 2018), direct transfection with plasmids containing DNA and the CMV promoter (Tsetsarkin et al. 2016; Schwarz et al. 2016), multi-fragment in vitro ligation transcription from the T7 promoter (Weger-Lucarelli et al. 2017; Widman et al. 2017; Deng et al. 2017), and multi-fragment transcription in cell recombination from the CMV promoter (Gadea et al. 2016; Atieh et al. 2016; Kobayashi et al. 2017). In these reversegenetic system platforms, reporter viruses are generated that either expressed Renilla luciferase (Rluc) (Shan et al. 2016; Münster et al. 2018), turbo far-red fluorescent protein FP635 (Münster et al. 2018) or GFP (Gadea et al. 2016). However, reporter Zika viruses are unstable after a limited number of passages on normal susceptible cells. Gadea et al. (2016) reported that recombinant ZIKV virus that expressed GFP lost the eGFP gene from the third passage on Vero cells. Reporter Zika virus expressing the RLuc gene was also unstable when passaged on Huh7 cells and Rluc activity steadily decreased with each passage and was lost after passage three (Münster et al. 2018). Moreover, reporter-gene insertion attenuated the replication efficiency of recombinant virus in normal susceptible cells, resulted in a low virus titer and the absence of plaques (Shan et al. 2016; Münster et al. 2018). Therefore, for studies involving live-cell imaging microscopy and/or high-throughput screening assays, more stable and more efficient reporter Zika virus systems are urgently required.
This study reports the development of a ZIKV reversegenetic system using homologous recombination methods. A stable ZIKV GFP-reporter virus system with considerably improved GFP visibility was developed, based on the genome sequence of Brazil epidemic strain ZIKV (Paraiba_01/2015). Using this reporter virus system, we established a high-throughput screening assay and screened a selected plant-sourced compounds library (TargetMol, Catalog No. L4600) for their ability to block ZIKV infection. Four compounds were identified that effectively inhibited ZIKV infection in cells. Two of these compounds are FDA-approved drugs, and here we report the anti-flaviviral activity of these two drugs for the first time. The active compounds identified in this study represent potential therapies for the treatment of ZIKV infection.
-
Human Embryo Kidney (HEK)-293T cells (ATCC, CRL- 3216), Vero cells (ATCC, CCL-81), and BHK-21 cells (ATCC, CCL-10) were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco C11995500CP) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, 10099-141), 100 U/mL penicillin and 100 lg/mL streptomycin, at 37 ℃ in 5% CO2. For HEK-293 T cell cultures, cell culture plates were covered with poly-D-lysine hydrobromide (Sigma P7886) at 50 lg/mL. The BHK-DR cell line (generated from BHK-21 cells as parental cells in this study), which stably expresses the dendritic cell-specific ICAM-3 grabbing non-integrin related protein (DC-SIGNR) that as a cell receptor may enhance the efficiency of virus infection, was maintained in DMEM supplemented with 10% FBS, 100 U/mL penicillin, 100 lg/mL streptomycin and 1 mg/mL G418 at 37 ℃ in 5% CO2. Zika virus and reporter Zika virus generated in this study were propagated on Vero cells and BHK-DR cells, respectively, and were titrated on BHK-21 cells and BHK-DR cells, respectively. Rabbit polyclonal antibodies against Zika virus envelope protein were obtained from GeneTex (GTX133314). Alexa Fluor 680-conjugated donkey anti-mouse IgG antibodies were obtained from Thermofisher Scientific (A10038). IRDye 800CW-conjugated donkey anti-rabbit IgG antibodies were obtained from LI-COR (926-32213). Chloroquine (Sigma C6628) and 6-azauridine (Sigma A1882) were obtained from Sigma.
-
To rescue Zika virus via homologous recombination in mammalian cells, the Paraiba_01/2015 strain of ZIKV, a Brazil 2015 epidemic strain was chosen as the sequence reference (GenBank accession number: KX280026). The Paraiba_01/2015 strain of ZIKV was isolated in the state of Paraiba (Brazil) in 2015 from serum of a febrile female (Tsetsarkin et al. 2016). The DNA-based ZIKV subgenomic replicon plasmid pZIKVrepdCME was constructed using the strategy described previously (Li et al. 2017b). The fragment of the first quarter of the ZIKV genome containing 5'-UTR-C-prM-E-NS1134 was synthesized and cloned into vector pCAGneo (Hua et al. 2014b) via XhoI and NotI restriction sites and the sequence-verified constructs were named pZCME-NS1134. To generate a GFPexpressing reporter virus, the GFP-encoding sequence fused to sequences encoding the first 33 amino acids of the C protein and the FMDV 2A coding sequence were inserted into pZCME-NS1134 between the 5'-UTR and the C protein gene. This construct was named pZC38GFPCME-NS1134.
The pCAG-DC-SIGNR construct expressing DCSIGNR was constructed by cloning the synthesized DCSIGNR-coding sequence (GenBank accession number: AY042234) into the SacI and XhoI restriction sites of pCAGneo.
-
To generate recombinant virus, the DNA fragments containing the CMV promoter and coding sequence for structural proteins were amplified by PCR from plasmids pZCME-NS1134 or pZC38GFP-CME-NS1134. For both plasmids, the PCR primers were: 5'-GGCCTTTTGCTCACATGGCTCGACAG-3' and 5'- GACTGCTGCTGCCAATCTACGGGGG-3'. The replicon plasmid pZIKVrepdCME was linearized using XhoI. The PCR products and linearized plasmids were purified using a DNA purification Kit (Omega, D2500-01). HEK-293T cells (90% confluent), were transfected with linearized plasmid and PCR-amplified DNA fragments using Lipofectamine LTX and PLUS Reagent (Invitrogen 15338-100). Three days following transfection, the supernatants were inoculated onto Vero and BHK-21 cells and the virus-infected cell cultures were harvested after 3 to 4 days. After three freezing-thaw cycles, the cell cultures were stored at - 80 ℃ for analysis.
-
The BHK-DR cell line stably expressing the DC-SIGNR protein was generated by transfection of BHK-21 cells with plasmid pCAG-DC-SIGNR as described previously (Hua et al. 2014a, b). Briefly, a monolayer of BHK-21 cells was transfected with the pCAG-DC-SIGNR plasmid using Lipofectamine LTX and PLUS Reagent (Invitrogen 15338-100). Two days later, transfected cells were digested and cloned by limited dilution in 96-well plates and grew in medium containing G418. The cloned cells were selected by indirect immunofluorescence assay (IFA) with mouse monoclonal antibody against DC-SIGN1/DC-SIGN2 (Sigma, D2691). The selected clone was designated BHKDR and maintained in G418-supplemented medium for further characterization and inoculation of ZIKV-GFP in high-throughput screening.
-
Virus-infected cells or BHK-DR cells were fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) at 4 ℃ for 10 min, and incubated with 4% bovine serum albumin V in PBS at 37 ℃ for 30 min. ZIKV was stained with pan-flavivirus E protein-specific mouse MAb 4G2 at 37 ℃ for 1 h or 4 ℃ overnight, then incubated with Fluorescein-Conjugated AffiniPure Goat Anti-Mouse IgG (ZSGB-BIO, ZF-0312) and Hoechst 33342 (Thermo 62249). To detect the expression of DCSIGNR, the primary antibody was a mouse monoclonal antibody against DC-SIGN1/DC-SIGN2 (Sigma, D2691). After three washes with PBS, cells were visualized using a fluorescence microscope (Life, EVOSFL).
-
Approximately 2 × 105 cells/well BHK-21 cells (for rcZIKV) or BHK-DCR cells (for rcZIKV-GFP) in 0.5 mL DMEM medium supplemented with 5% heat-inactivated FBS were seeded in 24-well plates, and 24 h later, the monolayers were inoculated with 100 μL of serially diluted virus. The viruses were diluted with DMEM medium without FBS. After 6 h, 0.6 mL 3% methyl cellulose (Sigma M6385) was added to each well. At 6 days after infection, supernatants were discarded and cells were stained with 1 mL crystal violet-formaldehyde stain for 15–20 min. Plates were washed twice with water and were air-dried. The resulting plaques were counted and plaqueforming units per mL were calculated.
-
Protein samples were separated by 12.5% SDS-PAGE, transferred onto a polyvinylidene difluoride (PVDF) membrane. To visualize proteins by immunoblotting, the following were used as primary antibodies: rabbit polyclonal antibodies against ZIKV E protein (GeneTex, GTX133314), mouse monoclonal antibody against GAPDH (Proteintech, 60004-1-Ig), rabbit polyclonal antibodies against GAPDH (Proteintech, 10494-1-AP). Primary antibodies were detected using Alexa Fluor 680 Donkey Anti-Mouse IgG secondary antibodies (Thermofisher Scientific, A10038) or IRDye 800CW Donkey Anti-Rabbit IgG (LI-COR 926-32213). Following four washes with PBS, signal detection was performed using a near-infrared fluorescence scanning imaging system (Licor Odyssey).
-
Approximately 3 × 104 cells/well BHK-DR cells in 100 μL DMEM medium supplemented with 5% heat-inactivated FBS were seeded onto CellCarrier-96 microplates (catalog number 6005550; PerkinElmer). After incubating the cells at 37 ℃ for 16 h, 50 μL dilutions of compounds were added to the cells. Approximately 1 h after addition of the compounds, 50 μL ZIKV-GFP was added to each well. Three wells of mock-infected cells and three wells of ZIKV-GFP-infected vehicle-treated cells were used on each plate as controls. The infected cells were incubated for 48 h at 37 ℃. Cell nuclei were stained with Hoechst 33342 (Thermo 62249). Infected cells were visualized using a high-content-screening microplate imaging reader (Operetta; PerkinElmer). Fifty-nine fields per well were imaged using a 209 objective. The images were analyzed using Acapella high-content imaging and analysis software. The infection-positive cell rates and total cell counts were all normalized to those of vehicle-treated control wells.
-
A library of 974 selected plant-sourced compounds was purchased from TargetMol (L4600). The compounds were stored as 10 mmol/L stock solutions in DMSO at - 80 ℃ until use. The first round of high-throughput screening was performed at a concentration of 10 μmol/L for each compound in triplicate. The criteria used to identify the primary candidates were a mean level of inhibition of ≥ 80% and relative cell viability of ≥ 90%.
-
Compounds selected in the primary screens were further evaluated by the same protocol as the primary screen, except that the compounds were serially diluted to final concentrations of 100 μmol/L, 10 μmol/L, 4 μmol/L, 2 μmol/L, 1 μmol/L, 0.5 μmol/L, 0.25 μmol/L, and 0.1 μmol/L. The 50% inhibition concentration (IC50) and 50% cytotoxicity concentration (CC50) were estimated using a nonlinear regression model from GraphPad Prism 5 software. The ability of four compounds with an IC50- < 1 μmol/L to inhibit viral replication was further evaluated in BHK-21 and A549 cells with wild-type recombinant ZIKV. One-hour following treatment with compounds at concentration of 8 μmol/L, 4 μmol/L, 2 μmol/L, 1 μmol/L, 0.5 μmol/L and 0 μmol/L, cells were infected with ZIKV at an MOI of 0.01. At 48 hpi, The ZIKV E protein expression level was determined and the amount of virus in the supernatant was titered to evaluate the inhibition efficiency of the compounds.
-
To evaluate which stage of the ZIKV life cycle was inhibited by each treatment, a time-of-addition experiment was performed as previously described (Chen et al. 2017; Aoki-Utsubo et al. 2018). Briefly, BHK-21 cells were infected ZIKV for 1 h (0 to 1 h). The test compounds were incubated with the cells or virus for 1 h before infection (- 1 to 0 h), during infection (0 to 1 h), and post-infection (1 to 48 h). At 48 h post infection, the supernatants were titrated and the cell lysates were immunoblotted to detect the viral protein expression level.
-
All statistical analyses were accomplished using GraphPad Prism 5. The P value was calculated using an unpaired Student's t-test. P > 0.05 was considered non-statistically significant (n.s.), and P < 0.05 was considered statistically significant, marked as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Cells, Viruses, and Antibodies
Plasmid Construction
DNA Transfection and Virus Production
Establishment of DC-SIGNR-Expressing Stable Cell Lines
Immunofluorescence Assay (IFA)
Plaque-forming Unit Assay
SDS-PAGE and Western blotting
Antiviral Screening Assay
High-throughput Screening of A Selected Plantsourced Compound Library
Compound Validation Studies
Time-of-addition Experiment
Statistical Analysis
-
To produce recombinant ZIKV, we first constructed a DNA-based replicon system pZIKVrepdCME (Fig. 1A). The first quarter of the ZIKV genome, containing 5'-UTRC-prM-E-NS1134, was synthesized and cloned into the pCAGneo vector under transcriptional control of the CMV promoter and resulted in pZCME-NS1134. To generate the GFP-expressing ZIKV reporter, plasmid pC38GFP-CMENS1134 was derived from pZCME-NS1134, in which the GFP reporter gene followed by the sequence encoding FMDV 2A was cloned in-frame downstream of sequences encoding the additional first 38 amino acids of the C protein and upstream of the complete C protein (Fig. 1A). The 2A protease ensured the authentic N-terminus of the C protein. DNA fragment A, containing the CMV promoter and 5'-UTR-C-prM-E-NS1134 of ZIKV, was amplified by PCR and was then used, in addition to linearized replicon plasmids, to transfect 293T cells (Fig. 1B). And DNA fragment B, which contained sequences of the CMV promoter and 5'-UTR-C38-GFP-2A-C-prM-E-NS1134, was amplified by PCR. Cells of 293T were transfected with linearized replicon plasmids and fragment B (Fig. 1B).
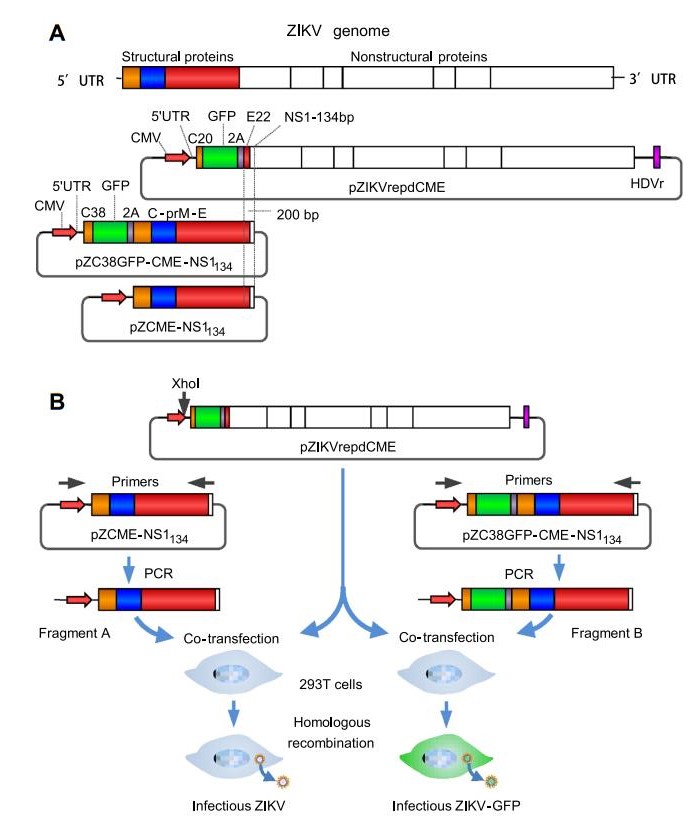
Figure 1. Schematic representation of the strategy for plasmid construction and the production of recombinant ZIKV and ZIKV-GFP. A Illustration of the ZIKV genome and constructs used in this study. In the pZIKVrepdCME construct, the GFP- and FMDV-2A-coding sequences were inserted downstream of sequences encoding the first 20 amino acids of the C gene and upstream of sequences encoding the last 22 amino acids of the E gene. The 30 untranslated region was flanked with HDVr to ensure the authenticity of the 30 terminus of the transcribed RNA. B Schematic diagrams for the production of recombinant ZIKV and ZIKV-GFP marker viruses. DNA fragments containing the CMV promoter and the coding sequences of structural proteins were amplified by PCR. The purified PCR products and enzymelinearized ZIKV replicon plasmid DNA were pooled and transfected into HEK-293T cells to generate infectious ZIKVs.
At 3 days post transfection, the supernatants were inoculated onto BHK-21 cells and infectious viruses were detected using an immunofluorescence assay (IFA) with pan-flavivirus E protein-specific mouse MAb 4G2 (Fig. 2A). The recombinant ZIKV was cytopathic and formed plaques in BHK-21 cells (Fig. 2B) and Vero cells (data not shown). Recombinant ZIKV grown on BHK-21 cells reached an infectious titer up to 5.7 log PFU/mL at 72 h post-infection (hpi) and then the virus titer decreased at 96 hpi and 120 hpi. Viral growth dynamics were then analyzed on Vero cells and the virus titer reached 6.7 log PFU/mL at 48 hpi and was maintained at 7.0 log PFU/mL from 72 to 120 hpi (Fig. 2C). These results showed that the replication of recombinant ZIKV in Vero cells was highly efficient. For rescuing ZIKV-GFP, at 3 days post transfection, the supernatants were inoculated onto BHK-21 and Vero cells to detect the titer of infectious virus. Positive GFP-expressing BHK-21 cells (Fig. 2D) and Vero cells were observed following inoculation with supernatants of transfected 293T cells. The infection of Vero cells with the recombinant reporter virus ZIKV-GFP caused cytopathic effects and led to plaque foramtion (data not shown). The infectious titer of ZIKV-GFP grown in Vero cells reached 6.5 log TCID50/mL at 72 hpi (Fig. 2E). However, the GFP signal in infected Vero cells was mainly observed as distinct foci of green fluorescence signal, which did not conform to the shape of cells (Fig. 2F). This observation weakens the utility of of ZIKV-GFP as a reporter virus. In infected BHK-21 cells, the green fluorescence signal was evenly distributed throughout the cytoplasm and the cell outline was apparent (Fig. 2D and 2F). However, the replication of ZIKV-GFP in BHK-21 cells was attenuated compared with that of wild-type recombinant ZIKV growth in BHK-21 cells. In BHK-21 cells infected with ZIKVGFP, the green fluorescence foci spread very slowly (Supplementary Fig. S1) and ZIKV-GFP did not cause cytopathic effects in BHK-21 cells even after incubation for 6 dpi or longer (Fig. 2D).
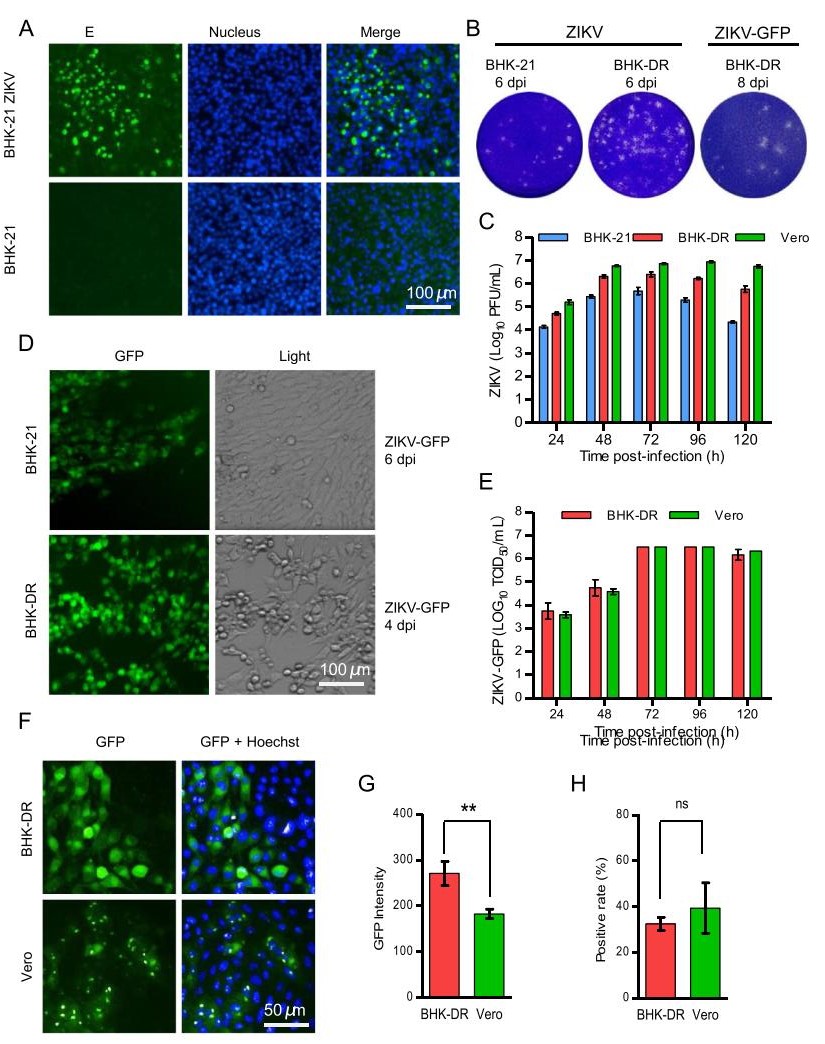
Figure 2. Production and characterization of recombinant ZIKV and ZIKV-GFP reporter viruses. A Immunofluorescence analysis of recombinant ZIKV. The supernatants of transfected 293T cells were inoculated into BHK-21 cells. Forty-eight hours later, infected cells were fixed and probed with pan-flavivirus E proteinspecific MAb 4G2. Nuclei were stained with DAPI. B Representative images of plaque assays of BHK-21 and/or BHK-DR cells infected with ZIKV and ZIKV-GFP viruses and fixed and stained at the indicated days postinfection. C Replication kinetics of recombinant wild-type ZIKV in BHK-21, Vero and BHK-DR cells. Cells were infected at an MOI of 0.1. Supernatants were harvested at 24 hpi to 120 hpi and analyzed using a plaque form assay in BHK-21 cells. D ZIKV-GFP reporter virus infection in BHK-21 and BHK-DR cells. Cells infected with ZIKV-GFP at the indicated time were analyzed by fluorescence microscopy for GFP expression and by light microscopy for cytopathic effects. E Replication kinetics of recombinant ZIKVGFP in Vero and BHK-DR cells. Cells were infected at an MOI of 0.1. Supernatants were harvest at 24 hpi to 120 hpi and analyzed using a TCID assay in BHK-DR cells by observation of fluorescent foci. F GFP fluorescence in Vero and BHK-DR cells infected with ZIKVGFP. One representative experiment out of three is shown. At 48 hpi, cells were stained with DAPI and visualized using a high-content screening microplate imaging reader (Operetta; PerkinElmer). The GFP fluorescence intensity G and GFP-positive ratio H were analyzed using Acapella high-content imaging and analysis software. The results are the mean of three independent experiments performed in triplicate. Statistical significance was determined by the Student's t-test (** P < 0.01, ns P > 0.05).
-
DC-SIGNR as receptor of flavivirus could increase the susceptibility of cells to ZIKV infection. To make ZIKVGFP a more readable and stable genetic reporting tool for viral replication studies and inhibitor screening, we decided to establish a cell line that stably expressed DC-SIGNR, to enhance the replication efficiency of ZIKV-GFP. Following transfection, selection with G418 and IFA identification, one clone was selected to further evaluate its ability to enhance the replication of ZIKV-GFP and was named BHK-DR. DC-SIGNR was expressed on cell membranes and in the cytoplasm (Supplementary Fig. S1). As expected, ZIKV-GFP replicated and the foci of GFP fluorescence spread more efficiently in BHK-DR cells than in BHK-21 cells (Supplementary Fig. S1). Infection with ZIKV-GFP caused cytopathic effects (Fig. 2D) and formed plaques (Fig. 2B) in BHK-DR cells. The virus titer reached a plateau at 6.5 log TCID50/mL at 72 hpi. The growth kinetic of ZIKV-GFP in BHK-DR cells was similar to that in Vero cells (Fig. 2E). We further assessed the infectivity of ZIKV-GFP in BHK-DR and Vero cells with a high-content screening system. Unlike the dot-like fluorescence observed in Vero cells, GFP fluorescence was observed throughout the cytoplasm of BHK-DR cells (Fig. 2F) and the cell outline was clearly identifiable. The GFP fluorescence intensity in infected BHK-DR cells was significantly higher than that in Vero cells (Fig. 2G), but the frequency of GFP-positive cells did not differ significantly between Vero and BHK-DR cells (Fig. 2H). These results indicate that the BHK-DR cell line was suitable for the propagation and visualization of the ZIKV-GFP reporter virus.
-
To assess whether the recombinant ZIKV-GFP reporter virus and BHK-DR cell system could be applied to the high-throughput screening for inhibitors of ZIKV, a reported inhibitor of ZIKV, chloroquine (CQ) (Delvecchio et al. 2016; Li et al. 2017a), was tested for its ability to inhibit infection of BHK-DR cells by ZIKV-GFP. Treatment with CQ inhibited ZIKV-GFP replication in BHK-DR cells in a dose-dependent manner (Supplementary Fig. S2). The number of infected BHK-DR cells was clearly reduced by CQ at a concentration of 4 μmol/L (Supplementary Fig. S2A). This observation was confirmed by quantitatively analyzing the infection rate using Acapella highcontent imaging and analysis. The relative infectivity of cells treated with CQ at 4 μmol/L or a higher dose was significantly lower than that of vehicle-treated cells (Supplementary Fig. S2C). The calculated IC50 value of CQ was 3.96 μmol/L, which was similar to previously published results with diverse screening systems (Li et al. 2017a).
The high-throughput system to screen ZIKV-GFP and BHK-DR cells was further evaluated using another reported flavivirus inhibitor, 6-azauridine (6-Az) (Lo et al. 2003; Adcock et al. 2017). Infection by ZIKV-GFP was inhibited by 6-Az at an IC50 of 7.16 μmol/L (Supplementary Fig. S2F), which was relatively consistent with the previously reported IC50 of 11 μmol/L against WNV (Lo et al. 2003) and an IC50 of 3.18 μmol/L against ZIKV strain MR766 (Adcock et al. 2017). A 6-Az concentration of 4 μmol/L significantly decreased the virus infectivity of treated cells (P < 0.001) compared with that of vehicletreated cells (Supplementary Fig. S2G). These results demonstrate that the HTS assay with ZIKV-GFP and BHKDR cells was effective and reliable.
-
To discover novel anti-ZIKV compounds or to repurpose approved drugs, we screened a library of 974 selected plant-sourced compounds for anti-ZIKV activity using a high-throughput system schematically depicted in Fig. 3A. The first round of screening was conducted at a compound concentration of 10 μmol/L. At this concentration, most compounds showed little or no inhibition effect on ZIKV infection. The subset of compounds with a relative infection inhibition rate from –40% to 40% accounted for 76.4% of the total (Fig. 3B). Cell viabilities were evaluated by normalizing cell counts to those of vehicle-treated cells. In total, 95 compounds showed a relative infection inhibition rate of > 80%. Among these 95 compounds, 31 (indicated by red circles in Fig. 3C) were selected for further evaluation. The 31 hits included dihydroartemisinin (DHA), a well-known anti-malarial drug, and homoharringtonine (HHT), a drug used to treat myeloid leukemia (Chen et al. 2009, 2019).
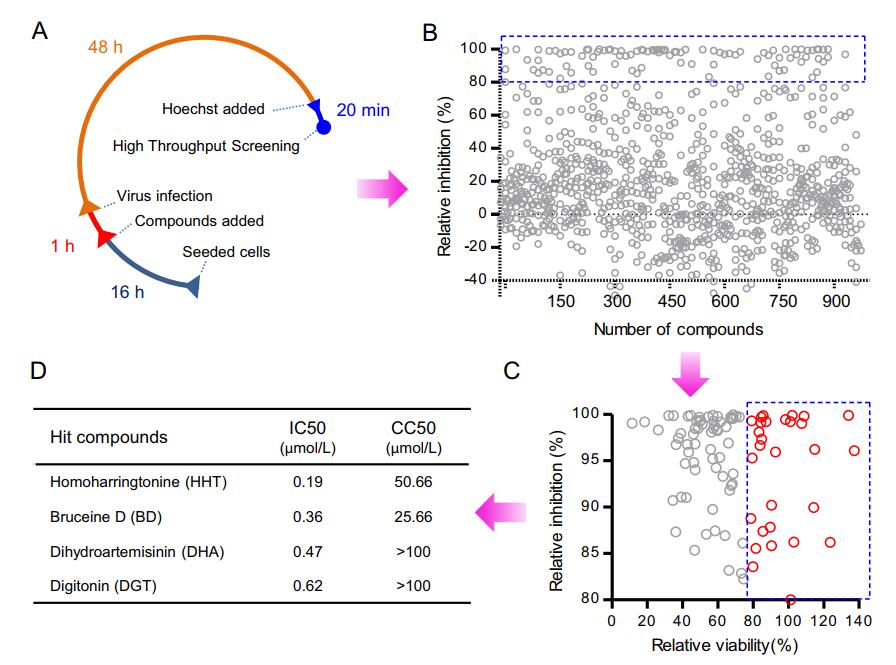
Figure 3. High-throughput screening for inhibitors of ZIKV infection from a selected plant-sourced compound library. A High-throughput screening (HTS) assay timeline. BHK-DR cells were seeded in CellCarrier-96 microplates. After incubation for 16 h (usually overnight), cells were treated with compounds. One hour later, cells were infected with ZIKV-GFP for 48 h. The final concentration of compounds was 10 μmol/L. The final concentration of vehicle (DMSO) was 1%. Cell nuclei were stained with Hoechst for 20 min. The plates were scanned using a high-content-screening microplate imaging reader. B High-throughput screening of a library of 974 selected plant-sourced compounds. Each circle represents the percentage inhibition achieved with each compound at a concentration of 10 μmol/L. The circles within the blue box represent an inhibition > 80%. In total, 103 compounds causing a virus infection inhibition rate of > 80% were selected for further analysis of relative cell viability. C Out of 103 compounds from the primary selection, 31 compounds represented by red hollow circles within the blue-dotted square passed the criterion of relative cell viability > 80% at a concentration of 10 μmol/L. These compounds were selected for a confirmatory screen. D IC50 and CC50 of the top four selected compounds.
-
In a primary screen, 31 compounds were selected for follow-up analysis based on the inhibition efficiency of ZIKV-GFP infection and the effect on cell counts. Each compound was used to pre-treat cells at concentrations of 0.1–100 μmol/L. As expected, most compounds displayed pronounced antiviral activity at a concentration of 10 μmol/L, which was consistent with the primary screen results. The selected 31 compounds included 19 with an IC50 ≤ 4 μmol/L (Table 1) and the most potent of these were homoharringtonine (HHT), bruceine D (BD), dihydroartemisinin (DHA) and digitonin (DGT) (Fig. 3D). All these compounds inhibited viral infectivity in a dosedependent manner (Fig. 4). The four compounds all possessed IC50 values less than 1 μmol/L. The virus infectivity of cells treated with HHT and BD at a concentration of 1 μmol/L was reduced by up to 90%; DHA and digitonin reduced virus infectivity by about 80%, without greatly affecting cell number, and showed CC50 values above 100 μmol/L (Fig. 4A). Cell number was significantly reduced by HHT and BD at a concentration of 100 μmol/L and the CC50 values were 50.66 and 25.66 μmol/L, respectively.
Compounds IC50 μmol/L CC50 μmol/L Homoharringtonine 0.19 50.66 Bruceine D 0.36 25.66 Cucurbitacin B 0.26 15.66 Dihydroartemisinin 0.47 > 100 Digitonin 0.62 > 100 Plumbagin 0.41 12.56 Schizandrin A 0.42 21.53 Dihydrochelerythrine 0.49 > 100 Betulonic acid 0.52 44.54 Artesunate 0.57 > 100 Sodium aescinate 0.69 > 100 Escin 0.76 > 100 Oleanonic acid 1.99 32.61 Cepharanthine 2.25 29.85 Corosolic acid 2.62 9.99 Bergamotine 2.98 23.76 Harringtonine 3.02 21.34 Schisanhenol 3.02 21.34 Fangchinoline 3.78 38.58 Fangchinoline 4.39 40.10 Dihydroactinidiolide 4.66 > 100 Schizandrin B 6.00 41.18 Tetrandrine 6.37 19.35 Liriope muscari baily saponins C 7.78 28.32 Ophiopogonin D 7.78 28.32 Saikosaponin A 9.22 27.72 Berbamine dihydrochloride 9.79 40.32 Daurisoline 10.13 29.28 Reserpine 10.20 33.33 Curcumin 11.23 > 100 Magnolol 31.37 > 100 After primary HTS, 31 compounds were selected for follow-up analysis. Each compound was used to pre-treat cells at concentrations of 0.1 μmol/L, 0.25 μmol/L, 0.5 μmol/L, 1 μmol/L, 2 μmol/L, 4 μmol/L, 10 μmol/L and 100 μmol/L. The vehicle was used as negative control. After 48 h of infection, cell nuclei were stained with Hoechst. Then the plates were scanned by using a high-contentscreening microplate imaging reader. Fifty nine fields per well were scanned using 209 objective and analysed for percentage of infection and cell number. The infection positive cell rates and total cell counts were all normalized to that of vehicle control wells. Table 1. Evaluation of the primary HTS selected compounds.
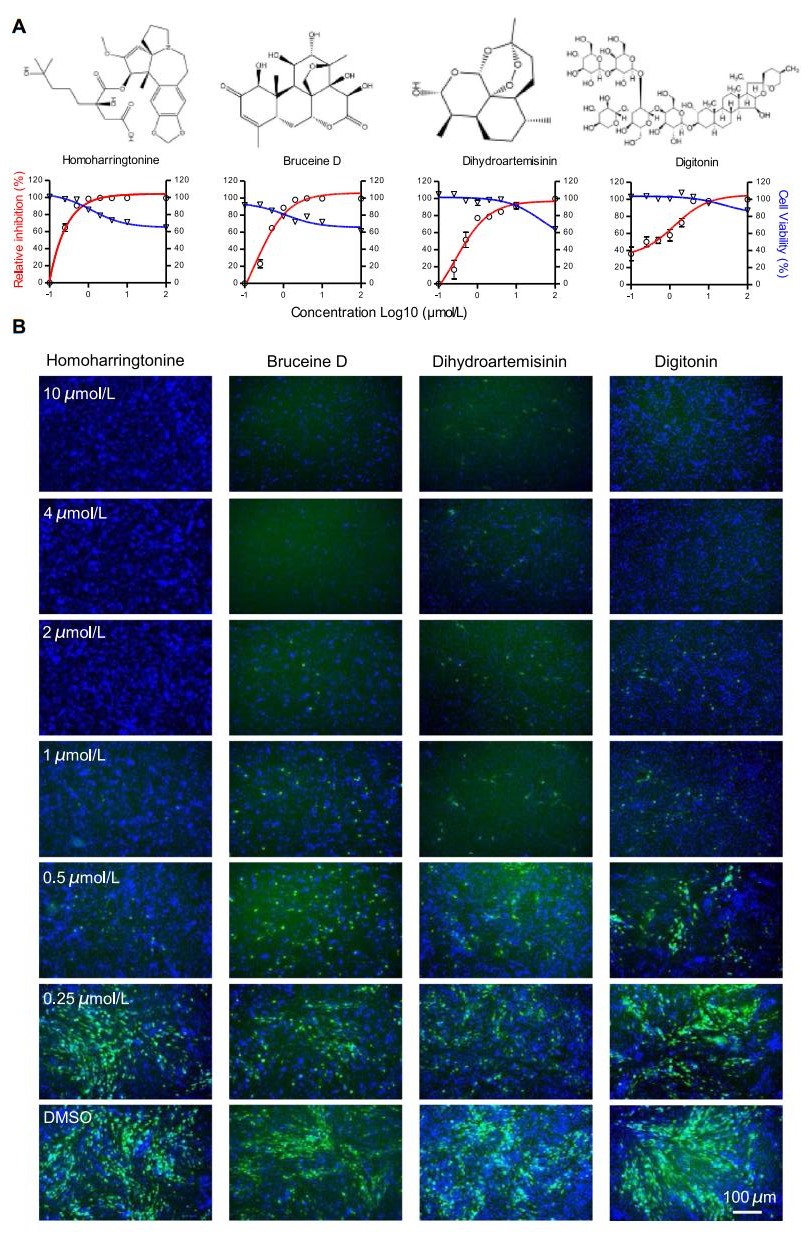
Figure 4. Validation of the antiviral effects of the hit compounds. A Top: The chemical structures of the hit compounds homoharringtonine (HHT), dihydroartemisinin (DHA), digitonin (DGT) and bruceine D (BD). Bottom: Dose–response curves showing the effect of compound treatment on virus infection inhibition (red) and cell viability (blue) in BHK-DR cells infected with ZIKV-GFP. Values represent the means ± SD from two independent experiments performed in triplicate. The data are normalized to those of DMSO-treated cells. B Representative fluorescence images of BHK-DR cells treated with varying concentrations of hit compounds. BHK-DR cells were treated with the indicated compounds at the indicated final concentrations and were infected after 1 h with reporter virus at an MOI of 0.1. Cells treated with DMSO at a final concentration of 1% were used as a vehicle control. At 48 hpi, cell nuclei were stained with Hoechst and cell plates were scanned using a high-content-screening microplate imaging reader.
The inhibition efficiency of the four selected compounds was further confirmed using BHK-21 cells and the human cell line A549 infected with recombinant wild-type Zika virus (Fig. 5). As expected, most compounds displayed inhibitory activity: 0.5 μmol/L HHT inhibited the E protein level by over 90% in both BHK-21 and A549 cells (Fig. 5A, 5E). For BD, ZIKV E protein levels in both cell types were significantly decreased at concentrations of 0.5 μmol/L (Fig. 5B, 5F). Treatment with BHK-21 cells at a DGT concentration of 8 μmol/L and A549 cells with 2 μmol/L DGT significantly inhibited the expression of ZIKV E protein (Fig. 5C, 5G). DHA treatment efficiently reduced the ZIKV E protein level in BHK-21 cells but only slightly inhibited that in A549 cells (Fig. 5D, 5H).
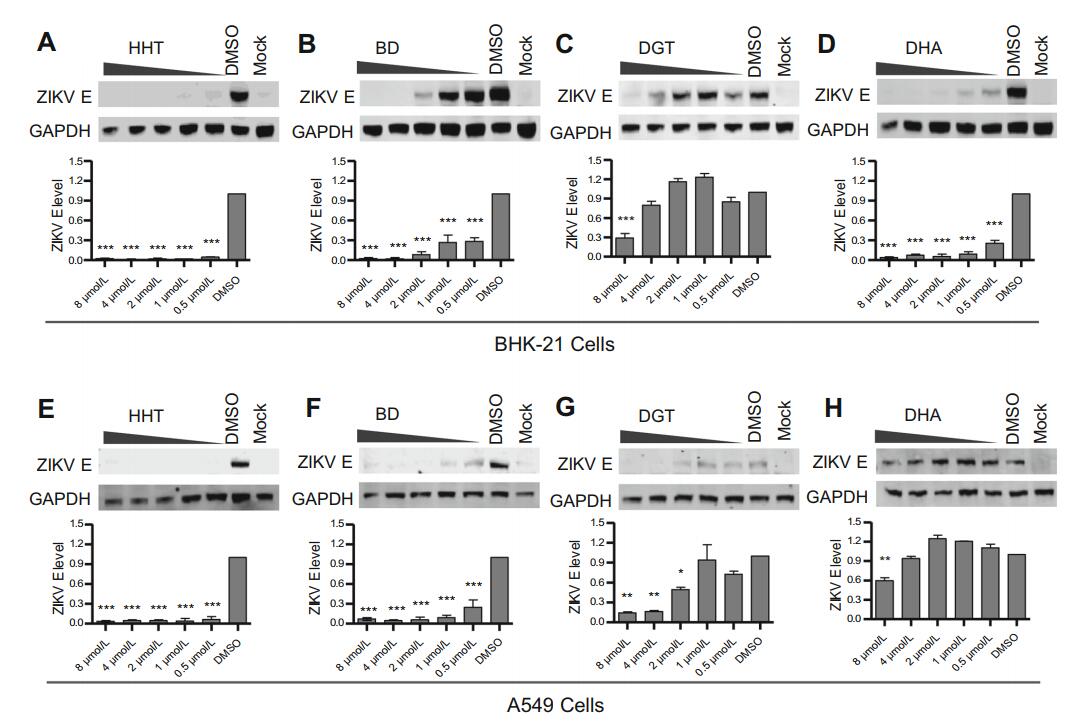
Figure 5. Confirmation of the inhibitory effect of selected compounds on ZIKV in BHK-21 and A549 cells. Each compound was used to pretreat indicated cells at concentrations of 8 μmol/L, 4 μmol/L, 2 μmol/L, 1 μmol/L and 0.5 μmol/L for 1 h prior to infection with wild-type ZIKV at an MOI of 0.01. Vehicle-treated cells and uninfected cells were used as controls. At 48 hpi, the cell lysates were analyzed by immunoblotting for ZIKV E protein expression, and the virus titer in the culture supernatants was measured on BHK-21 cells. BHK-21 cells treated with A HHT, B BD, C DGT and D DHA. Top: Representative western blot images. Bottom: Quantification of E protein band intensities relative to those for GAPDH. Data were normalized to those of DMSO-treatment cells. The data were pooled from two independent experiments. Values represent the mean ± SD. Statistical significances were determined by one-way ANOVA, compared to DMSO-treated cells (***P < 0.001). A549 cells treated with E HHT, F BD, G DGT and H DHA. Top: Representative western blot images. Bottom: Quantification of E protein band intensities relative to those for GAPDH. Data were normalized to those of DMSO-treatment cells. The data were pooled from two independent experiments. Values represent the mean ± SD. Statistical significances were determined by one-way ANOVA compared to DMSO-treated cells (*P < 0.05; **P < 0.01; ***P < 0.001).
-
To investigate the underlying cellular mechanism of the hit compounds that inhibited ZIKV replication, time-of-addition experiments were performed (Fig. 6A). The HHT (Fig. 6B), BD (Fig. 6C) and DHA (Fig. 6D) compounds all inhibited ZIKV infection at the post-entry stage, but not at the pre-entry stage or during infection. However, DGT reduced ZIKV E protein expression at all stages, including virus pretreatment, cells pretreatment, during infection and at the post-entry stage of ZIKV (Fig. 6E). Digitonin is a natural plant detergent, which affects the membrane proteins of viruses or cells and might inhibit the binding of viruses or their entry into host cells. Therefore, DGT exhibits an inhibitory effect at the early stage of ZIKV infection. This hypothesis requires further testing.
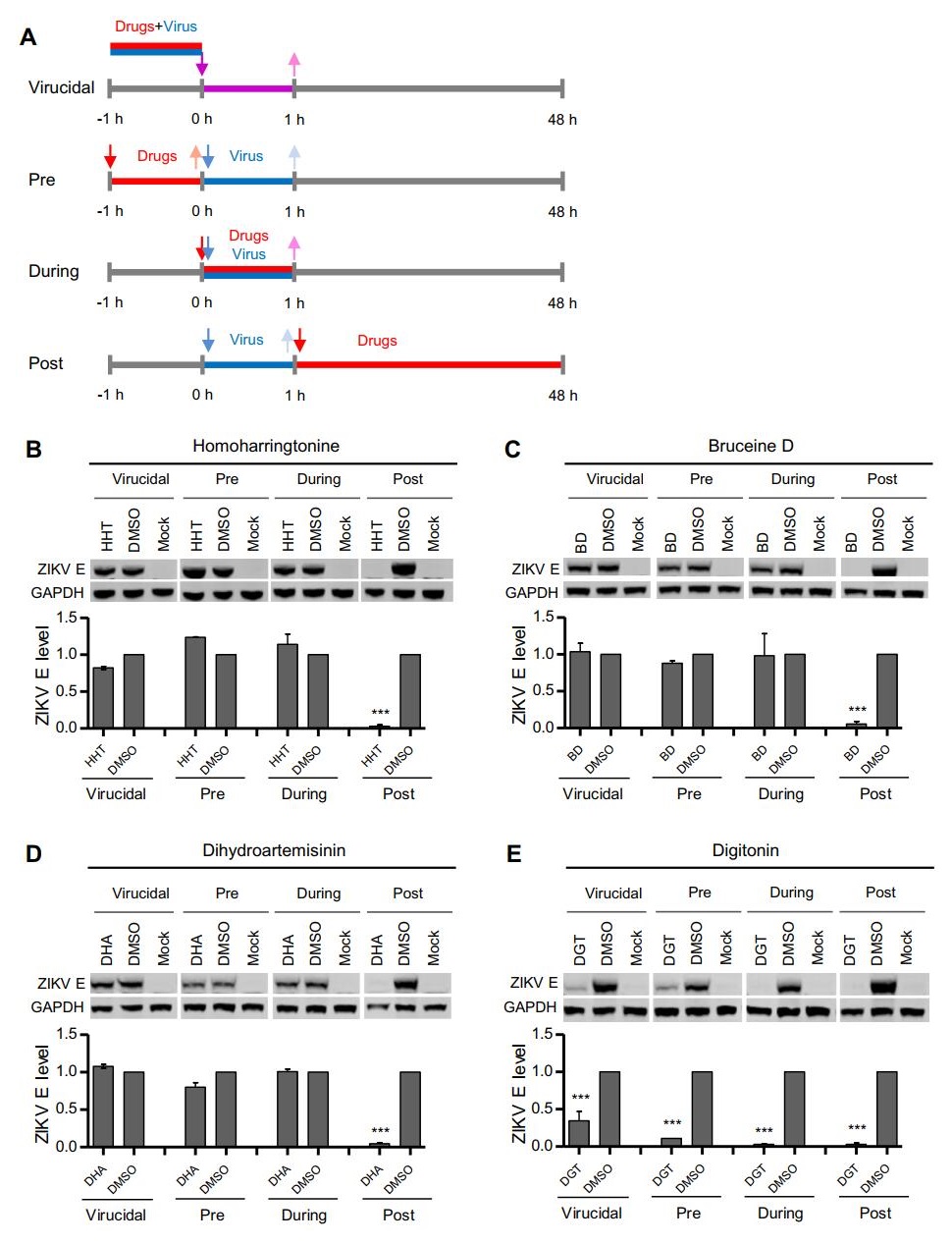
Figure 6. Time-of-addition analysis of the antiviral activity of the hit compounds. A Schematic illustration of the time-of-addition experiment. B, C, D and E BHK-21 cells were infected with ZIKV at an MOI of 0.01 for 1 h (0 to 1 h). Either 1 μmol/L HHT (B), 4 μmol/L BD (C), 4 μmol/L DHA (D), or 8 μmol/L DGT (E) were introduced at different time points of ZIKV infection, designated virucidal, pretreatment (pre), during treatment (during), or posttreatment (post). The inhibitory effect of the compounds in each group was determined by immunoblotting for E protein expression. Top: Representative western blot images. Bottom: Quantification of E protein band intensities relative to those for GAPDH. Data were normalized to those of DMSO-treatment cells. The data were pooled from two independent experiments. Values represent the mean ± SD. Statistical significances were determined by one-way ANOVA compared to DMSO-treated cells (***P < 0.001).
Generation of Recombinant Zika Virus via Homologous Recombination
Generation of A Stable DC-SIGNR-expressing Cell Line BHK-DR for the Propagation of Recombinant ZIKV-GFP Reporter Virus
Recombinant Reporter Virus ZIKV-GFP can be Used for High-throughput Screening for ZIKV Inhibitors
Screening of A Selected Plant-sourced Compound Library for Inhibitors of ZIKV Infection
Validation of Hit Drugs
HHT, BD and DHA Inhibit ZIKV Infection at A Post-entry Stage, Whereas DGT Is Effective at A Pre-entry Stage
-
Reverse-genetic systems are powerful tools with which to study virus biology and pathogenesis. Using different strategies of reverse-genetic systems, the classical African lineage strain MR-766 (Gadea et al. 2016; Schwarz et al. 2016; Widman et al. 2017; Münster et al. 2018), Asian lineage strain FSS13025 (Shan et al. 2016; Yang et al. 2017) and recent outbreak epidemic strains (Tsetsarkin et al. 2016; Weger-Lucarelli et al. 2017; Widman et al. 2017; Deng et al. 2017) of ZIKV were generated. In this study, we generated a recombinant ZIKV of Brazil 2015 epidemic strain via a homologous recombination reverse-genetic strategy, which involved cloning the ZIKV genome in two steps. Firstly, one-quarter of the genome that contains the 5'UTR, structural protein-coding sequences and the partial NS1-coding sequence was cloned into a plasmid under transcriptional control of the CMV promoter. The remaining three-quarters of the genome was constructed as a replicon plasmid. This reverse-genetic system overcame the difficulty of constructing a stable full-length flavivirus cDNA clone and furthermore, it is convenient to directly transfect cells with PCR products and plasmid DNA without in vitro transcription. Our results and previous reports have shown that this strategy is also highly efficient (Aubry et al. 2014; Atieh et al. 2016). In addition, the method provides a flexible strategy for constructing reporter viruses that express different foreign genes (Gadea et al. 2016), because only the plasmid containing the first quarter of the viral genome and the foreign gene needs to be reconstituted.
The use of a reporter virus can visualize the infection and replication of viruses in cells and is a powerful tool for studying viral replication mechanisms and to screen for antiviral drugs. However, to date, all reporter Zika viruses, including the expression of a luciferase reporter virus (Shan et al. 2016; Münster et al. 2018) or GFP reporter virus (Gadea et al. 2016) were unstable and their replication in cells was attenuated. These shortcomings limit the use of reporter viruses in the high-throughput screening of drugs and the mechanism of attenuated replication of reporter viruses in cells remains unknown. We observed that the replication of ZIKV-GFP in cells is slower than that of wild-type ZIKV, and the amount of green fluorescent foci increased slowly (Supplementary Fig. S1). These observations suggest that ZIKV-GFP mainly spreads via cell-to-cell contact infection, and that cell-free infection is weak. We hypothesized that increasing the cell-free infection of ZIKV-GFP might promote the replication efficiency of ZIKV-GFP in cells. Therefore, to increase viral infectivity in this study, we established a DC-SIGNR stably expressed cell line, BHK-DR, to promote viral replication. Because DC-SIGNR is a putative receptor of ZIKV and other flaviviruses (Navarro-Sanchez et al. 2003; Davis et al. 2006; Fernandez-Garcia et al. 2009; Shimojima et al. 2014; Hamel et al. 2015), its expression was expected to improve the replication efficiency of ZIKV-GFP and indeed, this was the case. This increase in replication efficiency potentially reduced the genetic stress of ZIKV-GFP. The reporter virus ZIKV-GFP remained stable in BHK-DR cells for at least five passages (data not shown).
Another potential reason for the observed increase in the replication efficiency and stability of the reporter virus is the number of amino acid residues of the C protein upstream of the foreign gene. We retained 38 amino acid residues of the C protein at the translation initiation. In fact, we also compared the effect of the retained length of C protein on the rescue of virus in this study. When only 28 residues were retained, the rescue efficiency of the reporter virus was lowest, few green fluorescent foci were observed and the reporter virus could be rescued occasionally. When 33 and 38 amino acid residues were retained, the virus could be rescued almost every time, and the retention of 38 residues led to the best replication efficiency (Supplementary Fig. S3).
To counter ZIKV-associated disease, efforts to develop effective anti-ZIKV drugs have increased worldwide and different platforms or systems have been established to screen for inhibitors of ZIKV. In addition to standard IFA and virus titration methods (Barrows et al. 2016), an assay with Renilla luciferase (Rluc) expressing the ZIKV replicon (Xie et al. 2016), the caspase-3 activity assay (Xu et al. 2016) and the Renilla luciferase reporter virus assay (Shan et al. 2016; Münster et al. 2018) have been developed to screen for anti-ZIKV drugs. Here, we have established a high-throughput screening platform for anti-ZIKV drugs using the BHK-DR cell line and the ZIKV-GFP reporter virus. In this system, the reporter virus is highly stable and has a high replication efficiency. Compared with the luciferase-expressing reporter virus screening system, our system does not necessitate the exchange of culture media or substrate addition and the results can be directly visualized and analyzed by live-cell imaging scanning.
Medicinal plants are a valuable source of therapeutic agents. Many of today's drugs originate from natural plant products or their derivatives, such as the anti-cancer agent paclitaxel and its derivatives (Kingston 2011; Cragg and Newman 2013) and the anti-malarial agent artemisinin (Atanasov et al. 2015). Out of the drugs considered to be basic and essential by the WHO, 11% originate exclusively from flowering plants (Veeresham 2012). Natural products remain a rich source of new antiviral leads, which might have broad-spectrum antiviral activities with novel mechanisms of action or improved resistance profiles. Using GFP expression as a read-out of Zika virus infectivity, we screened a selected plant-sourced compound library containing 974 compounds from 277 plant species. Homoharringtonine and bruceine D exhibited broad-spectrum inhibitory activity against ZIKV, Japanese encephalitis virus (JEV) and West Nile virus (WNV). Homoharringtonine is a natural product first discovered in the evergreen tree Cephalotaxus harringtonia, native to southern China. It has been used in China to treat chronic myeloid leukemia (CML), acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) for the past 40 years (Jin et al. 2006; Chen et al. 2019). Omacetaxine, a semisynthetic form of HHT, was approved by FDA in 2012 for the treatment of CML (Lü and Wang 2014). Besides the broad spectrum anti-flavivirus activities described here, HHT shows broad-spectrum inhibition of viruses from six families, including DNA and RNA viruses (Dong et al. 2018). Homoharringtonine has the potential to be repurposed as a broad-spectrum antiviral drug. Although the FDA lists it as a pregnancy Class D drug, it can be used in other infected individuals such as Guillain-Barré syndrome patients. Dihydroartemisinin is another FDA-approved antimalarial drug; however, its inhibitory effect against ZIKV in A549 cells was not as good as that in BHK21 cells and whether DHA can be used as an anti-ZIKV drug requires further evaluation. Bruceine D is a newly screened anti-ZIKV compound in this study. Its antiviral mechanism and its potential use as a drug remain to be further evaluated. Digitonin has inhibitory activity in the early stages of viral infection and is a non-ionic detergent, which also suggests that one positive method to prevent viral infections is via enhancing personal and environmental hygiene.
Most importantly, the data presented here describe an efficient and convenient high-throughput screening assay for ZIKV inhibitors, in addition to providing an additional approach towards anti-ZIKV drug development.
-
This work was partially supported by the National Key R & D Program of China (grant 2018YFC1200602 and 2016YFD0500403 to RHH).
-
RHH and ZGB designed the study. RHH, JWZ, HW, LM, JL performed the experiments. RHH and JWZ analyzed the data and drafted the manuscript. RHH and ZGB finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects peformed by any of the authors.
















 DownLoad:
DownLoad: