-
The interferons are a group of natural antiviral substances discovered in 1957 (7). Differential activeties of IFN subtypes have been reported (2). In particular, human interferon α2b, the most widely used member of IFN-α family, influences many biological processes including broad-spectrum antiviral effects, inhibition of tumor cell proliferation and enhancement of immune functions. The curative effect has led to the use of IFN-α2b as potential agents for therapy of neoplastic diseases, cancers and hepatitis (6, 8, 10, 15).
The powerful Pichia pastoris/pPICZα eukaryotic expression system, which can grow rapidly at high densities, was used to express the recombinant proteins at high-levels. The expression system carries the strong alcohol oxidase Ⅰ (AOX1) promoter, the mating factor signal sequence from S. cerevisiae and the Zeocin selectable marker (14). Although the final yield of a protein is greatly influenced by its inherent properties, the yield can be significantly enhanced by manipulation of the factors that influence gene expression and production stability (1).
To increase the expression level of IFN-α2b, the optimized strategies for enhancing the production level of proteins and synonymous codon usage bias in the P. pastoris expression system were applied in this study. Many studies have demonstrated that the use of P. pastoris synonymous codon usage bias in a variety of genes could result in high expression level and enhanced activity of the relevant gene products (12, 13, 16).
HTML
-
The coding sequence of IFN-α2b was designed by the authors and synthesized by Bio Basic Inc., Shanghai. Pfu DNA polymerase was purchased from Promega (Madison, WI) and all restriction enzymes was purchased from Sigma (Switzerland), EasySelectTM Pichia Expression Kit, was from Invitrogen (Carlsbad, CA), Mouse Anti-Human Interferon Alpha was from R&D Systems (Minneapolis, MN), AntiMouse IgG (Fc specific)-Peroxidase antibody produced in goat was from SIGMA (St. Louis).
-
The coding sequence of the synthetic IFN-α2b gene was amplified from plasmid pUC18M-IFN-α2b with primers IP1 (5'-GTGGAATTCATGTGTGACTTGCCACAAA-3') and IP2 (5'-ATAGCGGCCGCCTCCTTAGATCTCAAAGACT-3'), in which EcoR Ⅰ and Not Ⅰ site were introduced to 5' and 3' end, respectively. PCR was performed according to the following program: 1 cycle at 94 for 5 min, 30 cycles at 94 ℃ ℃ for 30 s, 60 for 30 s, 72 for 1 min, followed by 72 ℃ ℃ ℃ for 10min. The PCR products were purified and digested with EcoR Ⅰ and Not Ⅰ. The expression vector pPICZαA (Invitrogen) was also digested with EcoR Ⅰ and Not Ⅰ. All the digested fragments were ligated by T4 DNA ligase to yield the construct, designated as pPICZαA-IFN-α2b. The construct was transformed into TOP10F' and the positive colonies were identified by different restriction enzymes digestion and nucleo-tide sequencing. The positive colonies were then grown to prepare DNA for the following yeast transformations.
-
Pichia pastoris GS115 were grown in YPD medium and prepared for transformation according to the manufacturer's instructions. The pPICZαA-IFN-α2b was linearzed by Sac Ⅰ, and then transformed into P. pastoris GS115 by electroporation. Yeast transformants were incubated in YPDS medium containing 100μg/mL Zeocin at 30℃ for 2-3 d. A direct correlation between the number of integrated sequences and associated antibiotic resistance leads to an enrichment of the population of multi-copy strains (9). In order to screen out the multi-copy recombinants, the trans-formed yeast cells were picked on YPDS plates containing 0.5~2mg/mL Zeocin at 30℃ for 2 d (3). Multi-copy recombinants were selected, replica-plated onto minimal methanol (MM) and minimal dextrose (MD) agar plates and incubated at 30℃ for 2 d. Methanol utilization-plus transformants (Mut+) were selected by their normal growth on both MM and MD medium.
-
Each single colony was incubated with lyticase (Sigma) at 30℃ for 10 min, and then frozen at -80℃ for 10 min. The cDNA was isolated from the transformants as a template using the following primers: α-factor sequencing primer and 3' AOX1 sequencing standard primers. PCR was performed as follows: 1 cycle at 94℃ for 5 min, 30 cycles at 94℃ for 30 s, 65℃ for 30 s, and 72℃ for 1 min, followed by 72℃ for 10 min. The samples were analyzed by agarose gel electrophoresis. Each multi-copy recombinant was inoculated into 5 mL of cultural medium, BMGY (pH 6.0), and shaken vigorously (250 r/min) at 28~30℃ until OD600 reached 2~6. The cultures were centrifu-ged at 1500 g for 5 min at room temperature and the collected pellets were suspended in 50 mL of induction culture medium, BMMY (pH 6.0), until OD600 reached 1.0. The expression was induced at 28~30℃ with shaking (250 r/min) by addition of methanol every 24 h to achieve a final concentration of 0.5% (v/v). The supernatant of sampled was collected at various time points during 1~5 d to determine the optimal conditions.
-
The supernatant of the culture was collected at 10 000 g for 5 min, and mixed with SDS-PAGE Gel loading buffer. The supernatant was boiled for 10 min, then loaded onto 12% SDS-PAGE and electrophoresed for 1 h at 200 V. Another SDS-PAGE gel was transblotted onto a PVDF membrane (Millipore, Billerica, MA) for 1 h at 100 V. The membrane was washed and blocked with the blocking solution (TBS) containing 0.2% Tween 20 and incubated at room temperature for 2 h with the mouse anti-human interferon alpha as the primary antibody. The membrane was washed in TBS (containing 0.2% Tween 20) and incubated at room temperature for 1 h with anti-mouse IgG-peroxidase antibody. The membrane was washed again and visualized by ECL.
-
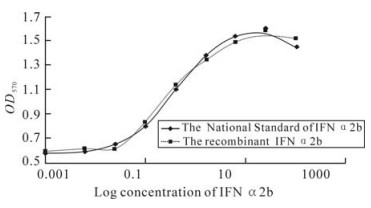
The antiviral activity of IFN-α2b was determined by their protective effect on human amnion WISH cells against Vesicular stomatitis virus (VSV) infection as described by the following Biological Products of China. In brief, 3.5×105 cells were seeded into each well of 96 well plates and incubated with four-fold serial dilutions of the culture supernatants of IFN-α2b and the National Standard of IFN-α2b respectively, for 24 h. The cells were then infected with VSV for 1-2 d. The survivor cells were visualized by staining with crystal violet and measured by optical absorption at 570 nm. The survivor cells show antiviral activity as 50% VSV infected cell with IFNα2b were alive.
-
Total amount of protein in the supernatant was assayed by the BCA method. Quantity of IFN-α2b was determined by scanning the area of each band on the SDS-PAGE gel and processing using the Quantity One software (Bio-Rad) (5).
Reagents and antigens
Construction of yeast expression vector
Yeast transformation, isolation of recombinants and determining Mut Phenotype
PCR analysis of pichia integrants and expression of recombinant protein
SDS-PAGE analysis and Western blot analysis
Antiviral assay
Analytical methods
-
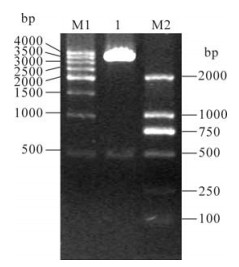
A synthetic IFN-α2b fragment on the basis of P. pastroris favored codons was generated (Fig. 1) (16). The synthetic fragment was designed to remove the native signal peptide sequence and had an optimal G+C content. After the synthetic IFN-α2b fragment was confirmed by sequencing analysis, the IFN-α2b fragment was amplified by PCR and digested with EcoR Ⅰ and Not Ⅰ, and inserted into the vector pPICZαA at the same sites to yield the expressing construct, pPICZαA-IFN-α2b. The construct was characterized by digesting with EcoR Ⅰ and Not Ⅰ into two fragments, one vector band of 3.6 kb and one target band of 512 bp (Fig. 2). These results indicated that pPICZαA-IFN-α2b was correctly constructed, which was further confirmed by sequencing analysis.
-
One thousand Zeocin-resistant colonies were generated through yeast transformation. Fifty-two multicopy integrants were selected on YPDS plates containing Zeocin at 0.5~2 mg/mL Zeocin. 43 transformants of Mut+ were isolated from these replicated Zeocin-resistant yeast colonies as identified by their normal growth on both MM and MD medium.
-
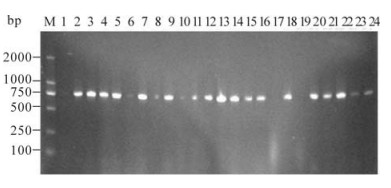
A DNA fragment of about 753 bp was amplified from transformed yeast cells by using α-factor sequencing and 3' AOX1 sequencing primers. The PCR amplification verified that the IFN-α2b gene had been integrated into the AOX1 locus on the chromosome of the transformed P. pastoris cell. Fortythree transformants were verified for the expression of pPICZαA-IFN-α2b gene product (Fig 3).
-
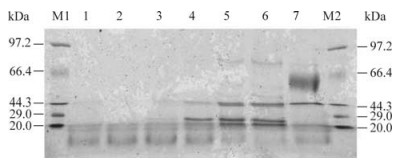
The transformants were induced to express IFN-α2b in small-scale testing with methanol in the 5 mL culture medium. After 96 h, the expressed proteins in the supernatants were detected by SDS-PAGE. A protein band (22.73 kDa) corresponding to the expected molecular weight of IFN-α2b protein was visualized on the gel. No similar band on negative control or blank control was observed. Clone B124 was selected to scale-up production of recombinant IFN-α2b. Protein expression of recombinant was induced for 0, 24, 48, 72, 96, 120 h and aliquots of 15 μL culture supernatant were detected by SDS-PAGE (Fig 4). Quantification of total protein was detected by the BCA Protein Assay. Quantitative estimation demonstrated that a maximum secretion of recombinant protein in excess of 810 mg/L was obtained at 120 h after induction with methanol.
-
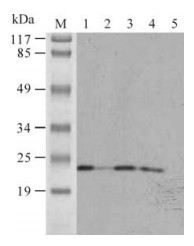
In order to test the reactivity of the recombinant IFN-α2b, western blotting was carried out and the protein band of 22.73 kDa was visualized with Mouse Anti-Human Interferon Alpha (Fig 5). These results confirmed the reactivity of the recombinant IFN-α2b.
-
The recombinant IFN-α2b protein showed signifi-cant antiviral activity. The antiviral activity was correlated with the dose of the recombinant protein and the relation between the antiviral activity and the dose of the recombinant protein showed a typical S-curve. Based on the National Standard of IFN-α2b, the antiviral activity of the recombinant IFN-α2b was3.3×105 IU/mL (Fig. 6).
Synthesis of gene for IFN-α2b and construction of yeast expression vector
Transformations and screening of multi-copy recombinants and determining Mut Phenotype
PCR screening of pichia clones
Expression of recombinant protein and SDS-PAGE analysis
Western blotting
Antiviral assay
-
To satisfy the requirement of high-level expression of the IFN-α2b for industrial production, this study has examined several optimized strategies that drasticcally influence expression of proteins in the P. pastoris expression system (12). First, the P. pastoris GS115 was chosen as the heterologous host to express IFN-α2b, as it could express foreign proteins at high levels extracellularly (4). In addition, the methanol inducible promoter (AOX1) played an important role in the high-level expression. Second, Secretion expression of IFN-α2b contributed to higher-level expression. Therefore, α-factor from S.cerevisiae replaced IFN-α2b signal peptide, which exported the recombinant protein IFN-α2b into the culture medium. Third, the special bias codons of P. pastoris were implemented in this study and the gene of IFN-α2b was modified according to the synonymous codon bias of P. pastoris and optimized G+C content, since optimizing codon sequence can improve expression level of heterogenous genes (16). Additionally, multi-ple-copy integration of recombinant genes in P. pastoris has been demonstrated in some cases to increase the expression of the desired protein, but multiple-copy integration occurred at a frequency of 1 to 10%. Resistance to higher levels of Zeocin is correlated with higher copy numbers of the integrated plasmid, which usually correlate with higher levels of expression of the recombinant target protein (11). Nearly one thousand Zeocin-resistant colonies were screened and fifty-two multiple-copy integrants were selected on YPDS plates containing Zeocin at 2 mg/ mL Zeocin. However, only 5 multi-copy integrants showed higher level of Interferon α2b expression by SDS-PAGE analysis. We presumed that the multicopy integrants have better site and mode of chromosomal integration of the expression cassette than other integrants.
The recombinant gene was also transformed into the SMD1168H in this study. However, the GS115 performed better in multi-copy clones of transformation and in recombinant protein expression than SMD1168H. We presumed that the SMD1168H without protease A activity competed against the transformation and the expression of the recombinant protein. Although the SMD1168 could avoid proteolysis of recombinant protein lacking of protease A activity, their effect was not effective in improving expression of the recombinant protein.
The expression level of IFN-α2b reached a maximum of 810 mg/L BMGY 120 h after induction with methanol using the optimized strategies. Therefore, the application of these strategies has greatly increased IFN-α2b expression. In this study, the levels of expression satisfies the goal of high-level expression of recombinant proteins in industry.














 DownLoad:
DownLoad: