-
Herpes simplex virus type 1 (HSV-1) is a doublestranded DNA virus with broad host range and mainly causes periodic and repetitive lesions of skins and mucosa. In addition, HSV-1 can present long-term latency in nerval cells or exhibit a potential carcinogenic capability. It is these characteristics that make the HSV-1 an important tool for conducting research in the fields of gene expression regulation, signal transmission, cell apoptosis and gene therapy.
HSV-1 gene expression in lytic infection proceeds in a tightly regulated temporal cascade. Six proteins, termed infected-cell polypeptide 0 (ICP0), ICP4, ICP22, ICP27, ICP47 and US1.5, have been identified as being expressed first. ICP0 has received the most attention due to its potent promiscuous transactivated capability on heterologous viral and cellular promoters in transient assays (3, 10). Moreover, ICP0 has been shown to interact with multiple cellular proteins such as ND10 nuclear structures(12), centomeric protein CENP-C (4), transcription factor BMAL1 (6); translation elongation factor EF-1δ(5), cyclin D3 (7), ubiquitin specific protease USP7(2) and Class Ⅱ histone deacetylases (8), highlighting its involvement in many aspects of host cell metabolism.
Compared to other HSV-1 proteins, expression of entire ICP0 in prokaryotic cells is very difficult--no such evidence has been presented so far. In this study, an antigen peptide of ICP0 was expressed with high efficiency in prokaryotic cells followed by preparation of the relevant antibody. Our results showed that such an antibody can not only recognize the denatured ICP0 protein in vitro, but also the native ICP0 protein in cells. This provides a useful tool for further investigation of its biological function.
HTML
-
The E.coli. DH 5α and BL21 (DE3) strains were purchased from Promega; the prokaryotic vector pGEX-5x-1 expressing glutathione S-transferase (GST) fusion protein was obtained from Pharmacia; the eukaryotic expression vector pcDNA3 was purchased from Invitrogen; the eukaryotic vector pDR27 expressing entire ICP0 cDNA sequences and pEGFP 110wt expressing ICP0 green fluorescence fusion protein were provided by Professors Peter O'Hare and Roger D Everett, respectively.
HeLa and Vero cells were grown in MEL medium supplemented with 10% bovine serum, 100U /mL penicillin and 20μg/mL streptomycin and cultured at 37℃ in a 5% CO2 incubator. The HSV-1 virus was separated from patient with herpes labialis in our laboratory and sequencing revealed a high homology with international standard stain (HSV-1 strain 17).
Various enzymes, PCR kits and IPTG were purchased from Takara Co.; LipofectamineTM2000 was from Invitrogen; Glutathione Sepharose 4B was from Pharmacia. Incomplete and complete Freund's adjuvant were from Sigma; ECL chemiluminescence reagent was from Beijing Tiangeng Biotech; the goat anti-mouse IgG with HRP and TRITC markers were from Beijing Dingguo Biotech; other reagents were imported or domestic products with high-grade purity.
-
Herpes simplex virus 1 with MOI of 0.05 was inoculated onto Vero cells washed in PBS, and then absorbed at 37℃ for 30 min followed by addition of MEL medium supplemented with 10% bovine serum. Cell cultures with obvious cytopathic effects were harvested and the dispersed cells were removed by centrifugation after 1 time freeze-thaw. VIVAFLOW 200 was employed for concentration of virus.
-
The forward primer P1 with EcoRI enzyme site (TATGAATTCATGGAGCCCCGCCCCGGA) and reverse primer P2 with XhoI enzyme site (TAACTCGAGGG GGGCCCCGCA) were designed by using plasmid pDR27 as template. cDNA sequence encoding the amino-terminal of 105 residues of the ICP0 protein was amplified by PCR. The PCR product was digested by the relevant enzymes and ligated into prokaryotic vector pGEX-5x-1 expressing the GST fusion protein for constructing recombinant plasmid pGEX-105. The amplification and transduction of plasmid were performed in E. coli DH 5α.
-
pGEX-105 plasmid was transformed into BL21 strain and incubated in 2xYT medium containing 100µg/mL ampicillin at 37℃ overnight. The cell cultures at 1:100 dilution were inoculated into 2×YT fresh medium containing 100µg/mL ampicillin and incubated at 37℃ until the OD600 reached 0.5. IPTG was added to a final concentration of 0.1mmol/L at 37℃ for 4 hrs to induce protein expression. The cells were collected after centrifuging for 10 min at 12 500r/min, and were immediately either identified by SDS-PAGE electrophoresis or frozen at –20℃. The BL21 strain expressing pGEX-5x-1 plasmid was used for controls.
In SDS-PAGE electrophoresis, the concentrated gel was 5% and separated gels were 12% or 8% based upon the molecular weights of proteins. Electrophoresis buffer contained 25 mmol/L Tris-Cl, 192 mmol/L glycine, 0.1% SDS, pH8.3. Staining was performed with Coomassie Brilliant Blue and decolorization with 10% ice acetic acid for autoradiography after electrophoresis, .
-
Induced E.coli cells were suspended in lysis buffer (containing 1g/L Triton X-100, PBS with 1 mmol/L PMSF, pH7.4) and dispersed in an ice bath by sonification until the suspension became clear. The lysate was then centrifuged for 15 min at 12 000g and 4℃. The supernatant was collected and mixed well with Glutathione Sepharose 4B prepared in PBS (pH7.4). The mixture was passed through an affinity chromatography column and washed by 5×volume of PBS (pH7.4) twice, and washed again by elution buffer (10 mmol/L reducting Glutathione, 50mmol/L Tris-HCl, pH8.0). After leaving at room temperature for 10 min the eluted volume was collected. The GST protein was obtained by the same approach. Protein content was determined by Lowry assay after being identified by SDS-PAGE electrophoresis.
-
The purified GST protein or GST-105 fusion antigen was mixed well with a ratio of 1:1 with the Freund's complete adjuvant and hypodermic injected into Kunming mice with an age of 6 weeks. The Freund's incomplete adjuvant emulsificated antigen was used to immunize the mice with a dosage of 50μg/mouse/time at week 2 and 4, respectively. The blood was taken from the optical fundus after 3 times immunization for serum preparation.
-
Monolayer HeLa cells grown to 95% confluent were transfected by LipofectamineTM2000. The nutrient medium was replaced by DMEM free serum before transfection. Plasmid was diluted in DMEM serum free medium and mixed with another serum free DMEM but containing LipofectamineTM 2000, and then incubated at room temperature for 20 min. The formed DNA-Lipofectamine 2000 complexes were added to cell medium dropwise. After 6 h, removed complexes and replaced by complete nutrient growth medium followed by another 42 h continuous culture. The HSV-1 viruses with MOI = 3 was used to infect the HeLa cells and cells were collected 18 h later.
-
Cells were treated with a detergent lysis buffer for SDS-PAGE electrophoresis. The protein was transferred to a nitrocellulose. 5% BSA was used to block the non-specific binding site and the membrane was bound with the obtained mouse serum at 1:500 dilution at 37℃. It was then washed 3 times with TTBS and incubated with rabbit anti-mouse IgG with HRP marker at 1:200 dilution at 37℃. The membrane was washed thoroughly and the protein was visualized by ECL assay.
-
The transfected HeLa cells were fixed in pre-cold 2% paraformaldehyde and washed in 0.2% Triton X-100/ PBS (pH 7.4) for 10 min. The 2% BSA/0.5% Tween20/PBS was used to block the non-specific binding site and bound to the obtained mouse serum at 1:500 dilution at 37℃. It was washed 3 times with PBS and incubated with goat anti-mouse IgG with TRITC marker at 1:100 dilution at 37℃. After another 3 washes in PBS, it was observed under a Nikon E600 fluorescence microscope.
Main reagents and materials
Culture and concentration of virus
Construction of recombinant plasmid pGEX-105
pGEX-105 expression in BL21 and SDS-PAGE analysis
Purification and quantitation of fusion protein
Preparation of antibody
Cell transfection and infection
Western blotting
Cell immunohistochemistry
-
The recombinant plasmid pGEX-105 was successfully constructed by enzyme digestion and DNA sequencing (Fig. 1).
-
After being induced for 4 hrs and identified by SDS-PAGE electrophoresis, proteins at about 26 kDa and 37 kDa were expressed in BL21 strain transformed by pGEX-5x-1 and pGEX-105 plasmids respectively, this is consistent with the GST and GST-105 protein of interest (see lane 4 and 5 in Fig. 2.). Further separation of supernatant and precipitation indicated that the concentration of GST-105 was very high in the supernatant of the lysed BL21 transformed by pGEX-105. High purity GST-105 fusion protein (see lane 6 in Fig. 2) was attained after Glutathione Sepharose 4B affinity chromatography purification; its concentration was about 378 μg/mL, as determined by Lowry assay. This protein could therefore be used to immunize the mice for preparing anti-serum. Meanwhile, the GST protein was used to immunize the mice as controls.
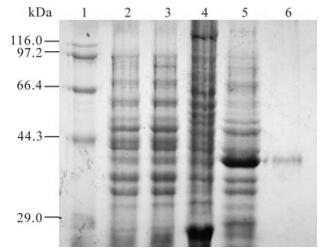
Figure 2. Expression and purification of ICP0 antigen peptide. 1, Marker; 2, BL21 (pGEX-5x-1) pre-induced; 3, BL21 (pGEX-105) pre-induced; 4, BL21 (pGEX-5x-1) 4h post induced by 0.1mmol/L IPTG; 5, BL21 (pGEX-105) 4h post induced by 0.1mmol/L IPTG; 6, Purified GST-105 protein (37 kDa) by Glutathione Sepharose 4B.
-
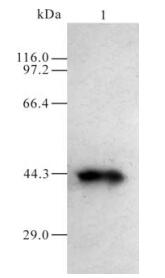
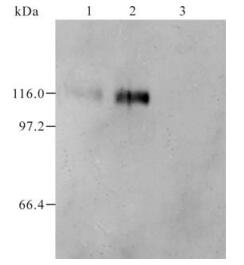
Serum obtained from the mice after immunization with GST and GST-105 was named as A0 and A105 respectively. A105 could specifically recognize the GST-105 protein of approximately 37 kDa (Fig. 3.). A band of about 110 kDa was recognized by A105 in the HeLa cells transfected by plasmid pDR27 and infected by HSV-1 (see lane 1 and 2 in Fig. 4.), whereas no band was recognized by A105 in the HeLa cells without pDR27 transfection and HSV-1 infection (see lane 3 in Fig. 4.). Furthermore, the reaction results of A0 in the HeLa cells transfected by pDR27 and infected by HSV-1 were negative (data not shown). Thus, these results reveal that the A105 antibody could specifically identify the denatured ICP0 protein in vitro.
-
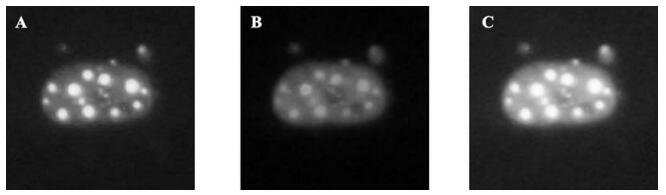
In order to further confirm that the A105 antibody could identify the native ICP0 protein in cells, we tried to use the A105 antibody to detect the ICP0 protein in HeLa cells transiently transfected with the pEGFP-110wt plasmid (encoding the GFP-ICP0 fusion protein). The results showed that typical dots of ICP0 protein were distributed in the nuclei, with exception of some diffused distributions. Moreover, the red fluorescence signal in the nuclei detected by A105 antibody were completely overlapped with the green fluorescence signal in nuclei detected by GFP-ICP0 (see Fig. 5.A-C), suggesting that A105 antibody could identify the ICP0 protein with a normal biological conformation.
Construction of recombinant plasmid pGEX-105
Expression and purification of GST-105 fusionprotein
Identification of antibody specificity
Native ICP0 protein identified by A105 antibody
-
ICP0 is an immediate-early protein playing a vital role in the replication of HSV-1. The complete encoding sequence of ICP0 is 2328bp in length and contains 3 exons. In recent years, more and more research has been focused on the regulatory functions of ICP0 on HSV and host cells. Conducting further studies on ICP0 will have an important theoretical and practical significance. However, no evidence of an entire ICP0 protein expression in prokaryotic cells has been reported so far. In our studies, different vectors (such as PET-28a, PGEX-5X-1, PBV-220 and PThio-His) carrying full-length ICP0 cDNA sequence failed to express ICP0 protein in different strains such as BL21(DE3) and Top10. Additionally, the constructed recombinant expression vectors encoding only the 3rd exon also failed to express in these strains. Yao and colleages reported that it is hard to get an entire ICP0 protein in prokaryotic cells, as this protein is discovered to be highly unstable in this environment (13). Our analysis reveals that the number of rare codons absent in E.coli accounts for 1/4 of the total codons in ICP0 and several rare codons are often continuously appeared, which might make it extremely difficult for an entire ICP0 to generated in E.coli. In light of such facts, we selected a cDNA fragment of ICP0 which contains relatively fewer numbers of rare codons and thus achieved the target of high expression with GST fusion protein. Since we only select a peptide of ICP0 as the immunogen, we should verify whether the attained antibody could identify the denatured ICP0 protein in vitro and the native ICP0 protein in cells as well. This interest was confirmed by the attained anti-serum A105.
Our studies also demonstrate some biological characteristics presented by the ICP0 protein. The theoretical molecular weight of ICP0 calculated by the Omega software is 78.46 kDa, whereas its apparent molecular size in the infectious process of virus is 110 kDa (thus it is termed IE110). Currently, such a fact is deemed as a consequence of the extensive modification after translation of ICP0, including the phosphorylated modification of the protein kinase UL13 of the HSV-1 and the cell cyclin protein Cdc2 (1, 11). In addition, the nucleosidation modification of casein kinase Ⅱ on ICP0 also plays a role (9). Our Western blot results indicate the onset of an apparent molecular weight of 110 KD in the absence of UL13 absence and by transfecting only the ICP0 eukaryotic expression vector. Similar results are reported by Zhu et al (14) who employ the recombinant adenovirus and Kawaguchi et al (7) who employ stable transfection strategies to obtain ICP0, this suggests some factors in host cells could replace the function of the protein kinase UL13.
The successful expression of the ICP0 antigen peptide in prokaryotic cells and the preparation of the ICP0 specific antibody in this report will help to further investigate the biological functions of ICP0 so as to provide advantageous strategies for preventing and controlling the infection of the HSV.













 DownLoad:
DownLoad: