-
Bluetongue (BT) is infectious and non-contagious, which is transmitted by haematophagous insects of the genus Culicoides, family Ceratopogonidae (4). BT has been included in the Office International des Epizooties (OIE) List of notifiable diseases (formerly List A) (12), and is a vector-borne animal disease of economical importance due to the international trade restrictions likely to be put into place in a country once the infection is discovered (14). Nations which are free from BT attempt to prevent the introduction and potential establishment of BT by imposing restrictions on the importation of hoofstock, including wild ruminants, from nations that either have the disease or share a common border with a country that has BT. BT occurred in South Africa in the late nineteeth century. At present, it has been distributed widely in over 50 nations of tropical, subtropical and temperate regions of the world. The disease is prevalent in 29 provincial areas of China (18). In July 2006 a sixth serotype, BTV serotype 8 was detected in Northern Europe (a new European serotype), with disease outbreaks in Belgium, Holland, Germany and France (5, 10), suggesting that the whole Europe is at risk from this and possibly other arbovirus diseases. In July and August 2007 bluetongue broke out again in Belgium, Spain, France and Netherlands (http://www.oie.int/).
Bluetongue virus (BTV), a member of genus Orbivirus, a family Reoviridae, and infects both domestic, captive and free-ranging ruminants. Twenty-five serotypes of BTV are distributed throughout the world (3). The virus is non-enveloped with a double shelled structure and an RNA genome consisting of 10 variously sized double-stranded (ds) RNA segments (8). The genome segments code for viral proteins (VP). Three nonstructural and seven structural proteins are incorporated into the double layer protein coat. The nonstructural proteins NSl, NS2 and NS3 are coded by genome segments 6, 8 and 10, respectively. VP7, which was reported to be a conserved protein, is coded by genome segment 7 (15). The NS1 gene of BTV is highly conserved among the 10 dsRNA of BTV serogroup (9).
Some improved methods based on PCR have been introduced which are important for BTV detection (1, 7, 11, 13, 17). However, these methods are inconvenient and are unable to rapidly characteristize or detect the virus in a universal manner. Therefore, development of a single set of novel primers for convenient and rapid detection of various BTV serotypes using conventional RT-PCR will facilitate the monitoring and prevention of the disease both effectively and rapidly.
To determine the conserved region, the VP7 genes (segment 7) of BTV 1, 2, 12, 16, 18 and 23, and the NS1 genes (segment 6) of BTV 10, 13, 17, 20 and 21 published on GenBank were aligned using the DNAStar Software package. Results revealed that the percent identities of VP7 genes ranged from 73.1% to 97.3%, while the percent identities of NS1 genes ranged from 79.0% to 98.7%, and the percent identity of the 5' end of NS1 gene containing a highly conserved region in the different serotypes was 93.8% (not shown).
According to the above results, the unique oligonucleotide primers were designed based on the 5' end of NS1 gene of BTVs (GenBank accession nos. AY462225, M97762, NC_006025, X15891, X56735 and Y00422). Primer 1 extended from position 10 through 30. Primer 2 was located from positions 110 through 130 of the complementary strand. These primers produced a 121bp PCR amplified fragments. The oligonucleotide sequences of the primers were as follows:
Primer 1: 5' to 3' GTTCTCTAGTTGGCAACCACC
Primer 2: 5' to 3' GTGACTGCAAGTCCATTGAGG
The sequences of the primers were searched for homologs in genes of other viruses published on GenBank (NCBI, http://www.ncbi.nlm.nih.gov/) using the Blast algorithm. The result revealed that the primers have homology to the BTV gene only (not shown).
To evaluate the practicability of the primers, BTV serotypes 1, 3, 5, 8, 10, 11, 21 and 22 reference strains (obtained from China Animal Health and Epidemiology Center) were assayed by RT-PCR using the above primers. PCR products were electrophoresed on a 2.5% agarose gel and observed by ethidium bromide-staining. The result as shown in Fig. 1 indicated that eight different serotypes of BTV were detected successfully.

Figure 1. Ethidium bromide-stained agarose gel electrophoresis showing the 121 bp PCR product from eight BTV serotypes, respectively. M, DL2000 marker; N, Negative control; 1-8, BTV 1, BTV 3, BTV 5, BTV 8, BTV 10, BTV 11, BTV 21 and BTV 22.
To confirm the specificity of the primers, BTV serotypes 1 to 12 and the closely related orbivirus Epizootic hemorrhagic disease virus (EHDV) serotypes 1 to 4 reference strains (obtained from Yunnan Entry-Exit Inspection and Quarantine Bureau) were detected with the above primers. The result showed that BTV serotypes 1 to 12 were detected by the primers as positive, whereas EHDV serotypes 1 to 4 were detected as negative (not shown). This indicated that the above primers are specific to BTV, and no cross reaction occurred with EHDV.
To prepare a standard sample pGEM-T121, the amplified products of BTV5 were cloned into pGEM-T Easy Vector (Promega, Madison, USA) and transformed into E. coli cells. The positive clones selected by blue-white screening and colony PCR were sequenced. Sequence analysis revealed that there was a perfect nucleotide match between the pGEM-T121 and detected fragment (not shown).
To estimate the sensitivity of RT-PCR with the primers, serially diluted pGEM-T121 were used as template and PCR were conducted using above primers. Result showed that the sensitivity of the assay with the primers was 105 copies/μL of the pGEM-T121 (equivalent to 3.4 × 10-4 μg/mL) (Fig. 2).
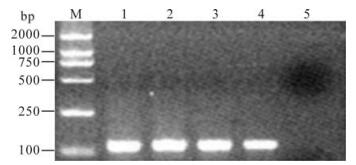
Figure 2. Sensitivity detection for pGEM-T121. M, DL2000 marker; 1-5, 1×108 to 1×104 copies of pGEM-T121.
To determine repeatability of the assay with the primers, the pGEM-T121 test was repeated three times, and five identical samples were tested each time. The results showed that all of the samples containing pGEM-T121 were positive.
To evaluate the actual value of the method with the primers, mimic specimens, which contained different concentrations of the re-suspended viral fluids and calf serum, were assayed using the primers. The mimic specimens were prepared by mixing 50μL, 100μL, 150μL and 200μL viral fluids of BTV 5 with 150μL, 100μL, 50μL and 0μL of BTV antibodies freed calf serum (GibcoTM, New York, USA) respectively. The mimic specimens were centrifuged at 8 000 r/min for 3 min, and the supernatants were subjected to viral RNA extraction and RT-PCR analysis using the above primers. The result con firmed that these samples could be detected as positive (Fig. 3).
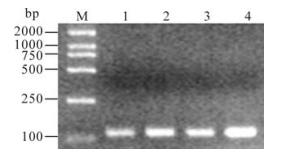
Figure 3. Detection of the mimic specimens with the assay. M, DL2000 Marker; 1, 50μL BTV5 + 150μL calf serum; 2, 100μL BTV5 + 100μL calf serum; 3, 150μL BTV5 + 50μL calf serum; 4, 200μL BTV5.
Monitoring and controlling of BTV infection in cattle, sheep and goats remain a top priority in BTVendemic and epidemic countries that are concerned with exporting livestock free of this disease or those restricting the introduction of new serotypes into already existing endemic populations. While serological assays (16) are sensitive and easy to use they only provide evidence of earlier animal exposure to BTV but not necessarily an ongoing infection. Earlier work on cattle and sheep has provided evidence that seropositivity does not correlate with circulating BTV RNA or the presence of infectious virus in the blood of the seropositive animals. PCR is the best-known and most successfully implemented diagnostic molecular technology to the date, and it can monitor the existence of virus particles (2, 6). Moreover, the technique is far simpler and much faster than a regular serological test and a viral isolation identification method. However, current RT-PCR methods with one pair of primers do not provide universal detection of different serotypes of BTV.
This study analyzed the conserved genes VP7 and NS1 of BTV. The highly conserved region 5' end of the NS1 gene was taken as the detected fragment and unique primers were designed based on this region. The results indicated that the novel primers could be used in RT-PCR to detect successfully and specifically for 14 different randomly selected serotypes of BTV, and no cross-reactivity with the closely related orbivirus EHDV was observed. The sensitivity of the specific RT-PCR was 105 copies/μL of the pGEMT121 in preliminary test, and this should be improved through optimization of reaction conditions. The repeatability of the method with the novel primers was good. Results also revealed that the mimic specimens could be detected successfully. Moreover, completion of the assay required approximately only 3 h. In this study, each experiment was performed in triplicate with identical results.
As a summary, the distinct primer set is suitable for detecting the NS1 gene of different BTV serotypes using RT-PCR, and has the potential to be a useful tool for high throughout screening and monitoring of BTV.
A Pair of Novel Primers for Universal Detection of the NS1 Gene from Various Bluetongue Virus Serotypes
- Received Date: 29 August 2007
- Accepted Date: 14 November 2007
Abstract: Twenty five serotypes of Bluetongue virus (BTV) have been identified worldwide. Rapid and reliable methods of virus universal detection are essential for fighting against bluetongue (BT). We have therefore developed and evaluated a pair of primers which can detect various serotypes of BTV by RT-PCR. Analysis of the viral protein 7 (VP7) and the non-structural protein (NS1) gene from different serotypes of BTV by DNAstar showed that the 5’ end of the NS1 gene is the most conserved region. The primer pairs (P1 and P2) were designed based on the highly conserved region of NS1. The novel primers were evaluated by detecting BTV serotypes 1, 3, 5, 8, 10, 11, 21 and 22. The specificity of the primers was estimated by comparing to gene sequences of viruses published in GenBank, and further assessed by detecting BTV serotype 1-12 and Epizootic hemorrhagic disease virus (EHDV) serotype 1-4. The sensitivity and repeatability of PCR with the novel primers were evaluated by successfully detecting the recombinant plasmid pGEM-T121 containing the diagnosed nucleotide sequence. Our results suggest that these unique primers can be used in high throughout and universal detection of the NS1 gene from various BTV serotypes.













 DownLoad:
DownLoad: