-
The global HIV epidemic continues to expand, exceeding previous predictions and causing tremen-dous suffering. It has been the leading cause of death in Africa. At the same time, a rapid increase of HIV infection has also been found in China in recent years. Therefore, developing HIV vaccines targeting the prevalent strains in China is one of the most important tasks for Chinese HIV/AIDS control and prevention.
Virus-specific cellular immune responses play an important role in the control of HIV infections (1, 6, 23). Recently some studies indicated that only Gag-specific responses were associated with lowering viremia in an untreated HIV-1 infected cohort and HIV-2 long-term nonprogressors, while Env-and Nef-specific responses are positively correlated with a high viral load (4, 7, 13, 15, 16). Thus Gag would be the preferred antigen in HIV candidate vaccines. DNA vaccines, recombinant-viral-vector based vaccines, and their combinations are promising AIDS vaccine methods because of their potential for inducing cellular immune responses. DNA vaccine is safe and easy to manufacture, and it is quite effective as a priming or initial immunogen in a bimodal vaccine strategy (17, 22); Some of the extraordinary features of the SeV vector are the remarkably brief contact time that is necessary for cellular uptake, a strong but adjustable expression of foreign genes, and an exclusively cytoplasmic replication cycle without any risk of chromosomal integration (5). An extremely efficient antigen expression system in mammalian cell cultures using recombinant SeV were established in the late 1990's (8, 12, 24). Using this system Kano et al demonstrated the excellent protective efficacy of DNA priming followed by simian immunodeficiency virus (SIV) Gag-expressing Sendai virus boosting against a pathogenic simian-human immunodeficiency virus (SHIV89.6PD) infection in macaques (10). Takeda et al obtained similar results by using a DNA prime/replication defective SIV Gag-expressing SeV boost system (19). In the present study a HIV-1 gag gene was used to construct DNA and replication defective rSeV vaccines. High-frequency CTL responses may be elicited by combining these two vaccines.
Although the discontinuation of the Merck rAd5 Phase Ⅱ proof-of-concept STEP study is undoubtedly a significant setback for the field of HIV vaccine development (18), it does not indicate the failure of the T-cell vaccine concept. In their study a trivalent recombinant adenovirus type 5 vaccine expressing Gag, Pol, and Nef respectively was tested. The issues of lack of vaccine efficacy and differential infection rates between vaccine and placebo groups with previous Ad5 immunity are complex ones and require careful consideration. Selection of T-cell vaccine antigens for chronic persistent viral infections has been largely empirical. Though immunization with more than one immunogen (co-immunization) is an efficient regimen to induce immunity to multiple antigens, Toapanta et al found that when HIV-1 Env (gp120) and HIV-1 Gag (p55) DNA plasmids were co-inoculated, there was a reduction in the immune responses elicited to HIV-1 Gag (p55). This anti-HIV-1 Gag immune interference was specific to co-immunizations with HIV-1 Env (gp120) and may involve a yet undefined immunological mechanism (20). Recently Kiepiela et al performed a compre-hensive analysis of the 160 dominant CD8+ T cell responses in 578 untreated HIV-infected individuals from KwaZulu-Natal, South Africa. They found only Gag-specific responses were associated with lowering viremia (13). So in our study Gag-expressing vaccines would be investigated first, and only if excellent immune efficacy could be induced would other antigens be included in a further study.
Most worldwide vaccine developments have been focused on inducing protection against prevalent strains in North America and Europe. Developing HIV vaccines targeting the prevalent strains in China is of great importance Thailand subtype B is found to be prevalent in several epidemic regions where paid blood donors are the principally affected population. The HIV-1 Subtype B in these epidemic regions is relatively well conserved, so the prevalent strains isolated from these regions was used as the vaccine strain. The constru-ction of DNA and rSeV vector vaccines containing the gag gene of prevalent Thailand B strains in China was therefore the main objectives of our studies.
HTML
-
The codon-modified consensus gag gene from HIV-1 prevalent strains in Henan province was used to construct a DNA vaccine. Firstly, the gag genes were amplified by nest-PCR using specific primers from the DNA from the peripheral blood mononuclear cells (PBMC) of HIV-1 infected patients in Henan province. Twelve gag sequences were obtained from 20 patients and 6 of them had complete open reading frames (ORF). The consensus gag gene was generated by sequence alignment of the 6 gag genes. The consensus gag sequence had different amino acids in 6 positions compared with reference sequences of subtype B (B.FR.HXB2-LAI-IIIB-BRU and B.TH. BK132). 95 % and 96 % of amino acid sequence homology were found with B.FR.HXB2-LAI-IIIB-BRU and B.TH.BK132 respectively. To increase the expression level of Gag protein in DNA vector the codons of the consensus gag gene were modified according to mammalian condon usage. The codon-modified gag gene was synthesized and inserted into plasmid pUC57 by the Shanghai Sangon Biological Engineering Technology & Service Co., Ltd (Shanghai, China). KpnⅠand XhoⅠrestriction enzyme sites were inserted into the gag gene by PCR and cloned into pcDNA3.1 (+) which was named pcDNA3.1 (+)-gag. The pcDNA3.1 (+)-gag was standardized at 1mg/mL in endotoxin-free Buffer TE.
The wild-type gag gene was used for construction of the recombinant Sendai virus containing gag (rSeV-gag). The rSeV-gag was constructed and amplified by the DNAVEC Corporation (Tsukuba, Japan) as follows: (ⅰ) EIS sequence and NotⅠ restric-tion enzyme sites were incorporated into the gag gene by PCR, cloned into pBluescript KS (+) and was named pBS-HIVgag. (ⅱ) pBS-HIVgag was digested with NotⅠ and the DNA fragment containing gag was inserted into pSeV/ΔF (a viral genomic RNA-en-coding plasmid) that contains SeV full-length cDNA lacking the F gene. The generated plasmid was named pSeV18+HIVgag/ΔF. (ⅲ) Recombinant virus con-taining the gag gene was recovered by co-transfection of 293T/17 cells with pCAGGS-P(z)/4C-(P protein-expressing plasmid), pCAGGS-NP (NP protein-expressing plasmid), pCAGGS-L(TDK) (L protein-expressing plasmid), pCAGGS-F5R (modified-F protein-expressing plasmid), pCAGGS-T7 (T7 RNA polymerase-expressing plasmid) and pSeV18+HIV gag/ΔF. (iv) Recovered virus was cloned and amplified in LLC-MK2/F/A cells expressing the F protein. Virus yield is expressed in cell infectious units (CIU). The recombinant Sendai virus was titrated as follows: LLC-MK2 cells were seeded into a 6-well plate at a cell density of 2×105 cells/2mL/well and incubated at37℃, 5%CO2 for 72 h until 100% confluence. 10-fold dilutions of the virus stock were prepared in PBS containing 1% BSA. The confluent monolayer was washed once with PBS and then was inoculated in duplicate with 0.1 mL of the virus samples/well. The cells were incubated for 1 h at 37℃, 5% CO2, and the plate was tilted every 15 min. Virus inoculum was removed, the cells were washed with PBS and supplemented with 2 mL of DMEM and incubated at 37℃, 5% CO2for 48h. The cells were washed with PBS and fixed with methanol for 10 min at 4℃. After removing methanol the plate was dried for 10 min at room temperature. 0.5 mL of 1:500 diluted polyclonal rabbit-anti SeV antibodies (prepared by the DNAVEC Corporation) was added into each well. The plate was incubated for 45 min at 37℃ then was washed twice with PBS. 0.5 mL of 1:200 diluted goat anti-rabbit immunoglobulin G (Zhongshan Goldenbridge Biotech-nology Co., LTD, Beijing, China)was added into each well. The plate was incubated for 45 min at 37℃ then was washed twice with PBS. The number of positive cells was counted under the fluorescence microscope. The virus titer was calculated by the formula: mean numbers of positive cells in duplicate wells × dilution multiple×10 (CIU/mL). The titer of generated rSeV-gag was 1.8 × 1010 CIU/mL.
-
LLC-MK2 cells were seeded into a 6-well plate at a cell density of 2×105 cells/2mL/well and incubated at 37℃, 5%CO2 for 72 h to 100% confluence. The confluent monolayer was washed with PBS, then was inoculated with 0.1 mL of the virus samples/well (MOI of 5, to count the cell number per well, the cells in one well was trypsinized and counted under a microscope). The cells were incubated for 1h at 37℃, 5% CO2, and the plate was tilted every 15 min. Virus inoculum was removed, the monolayer was washed with PBS and supplemented with 2mL of DMEM and incubated at 37℃, 5%CO2 for 48 h. The virus which did not contain the foreign gene (rSeV-control) was used as the control. 48 h later, total RNA was isolated from infected cells using Trizol reagent (Promega, Madison, USA). One-Step RT-PCR was used to test the presence of the gag gene in the recombinant virus (wt269F: 5'-GGCAAGCAGGGAACTAGAAC-3', wt-269R: 5' AGAACCGGTCTACATAGTCTC-3').
-
LLC-MK2 cells were maintained in DMEM and supplemented with 10% fetal bovine serum (FBS). Cells were transfected with pcDNA3.1(+)-gag or infected with rSeV-gag at a MOI of 5. Proteins of transfected or infected LLC-MK2 cells were extracted 48 h later using TRIzol reagent. The protein samples were subjected to SDS-PAGE and electroblotted onto nitrocellulose blotting membranes. Blots were blocked with 5% fat free milk in phosphate buffered saline (PBS) containing 0.05% Tween 20 and probed with mouse anti-HIV-1 P24 monoclonal antibody(NIH AIDS Research and Reference Reagent program, Germantown, USA)and peroxidase-conjugated goat anti-mouse immunoglobulin G (Zhongshan Goldenbridge Biotechnology Co., LTD, Beijing, China). Proteins were visualized by staining with 3, 3'-Diamino-benzidine.
-
Four to six-week-old Balb/c female mice were purchased from Institute of Experimental Animal Sciences, Chinese Academy of Medical Science (Beijing, China). To compare the immunogenicity of the DNA and rSeV-gag vaccines applied in single or combined vaccines, mice were inoculated with these vaccines intramuscularly either in single or combined modality at week 0 and week 3. The immunization dose for the DNA vaccine was 100μg per animal and for the rSeV vaccine was 1.8 ×107 CIU per animal. Balb/c mice were randomly divided into four groups of 16. Inoculation was conducted according to the schedule in Table 1. The splenocytes and sera of immunized mice were collected at 1, 5 and 9 weeks post immunization and the cellular and humoral immune responses were analyzed.

Table 1. Immunization schedule of DNA and rSeV vaccines
-
Freshly isolated splenic lymphocytes (2×106 cells) were suspended in 10% FBS RPMI1640 medium and incubated with H-2d-restricted CTL epitope peptides, Gag197-205(AMQMLKETI), Gag239-247(TTSTLQEQI) and Gag291-300(EPFRDYVDRF) for antigen-specific stimulation or without peptides for mock stimulation. The three peptides were pooled together and the final concentration for each peptide was 10µg/mL. Cells were cultured for 16h at 37℃. For the final 12 h, brefeldin A (Sigma, Saint Louis, USA) was added at 5μg/mL. After stimulation, the cells were stained (60 min, 25℃) for surface markers with 5 μL of R-PE-conjugated rat anti-mouse CD8a (Ly-2) monoclonal antibody (BD Pharmingen, San Diego, USA). Then the cells were sequentially fixed with 4% paraformal-dehyde and permeabilized with 0.3 % saponion for 15 min respectively, and stained for 60 min with 1 μL of FITC-conjugated rat anti-mouse IFN-γ monoclonal antibody (BD Pharmingen, San Diego, USA). Stained samples were collected by a Coulter EPICS Altra flow cytometer (Beckman, Fullerton, USA) and analyzed using the CellQuest software package. Gating was performed on mononuclear cells and then on CD8+ subpopulations. 50 000 of CD8+ T cells were collected in total. From the ratio of CD8+IFN-γ+ cells to CD8+ cells, the frequency of CD8+ IFN-γ+ cells in the total CD8+T cells was calculated. Gag-specific T-cell frequencies were calculated by subtracting the CD8+IFN-γ+-cell frequencies after mock stimulation from those after Gag-specific peptides stimulation.
-
Specific antibodies were detected by the enzyme-linked immunosorbant assay (ELISA). 96-well microtiter plates were coated with 200ng/well of HIV P24 protein prepared by our laboratory and incubated overnight at 4℃. The wells were blocked with PBS containing 5% fat free milk for 1h at 37℃. They were then treated with 100 μL of serially diluted mice sera and incu-bated for an additional 1h at 37℃. The plates were washed five times with PBS containing 0.05% Tween-20 and incubated for 1h with 1:20 000 diluted goat anti-mouse IgG/HRP. The plates were then washed five times, developed with tetramethylbenzidine, stopped with 2 mol/L H2SO4, and analyzed at A450nm/630nm.
Construction of DNA and rSeV vaccines
Identification of gag gene in rSeV-gag by RT-PCR
Identification of Gag expression in DNA and rSeV-gag vaccines
Animals and immunization
Intracellular IFN-γ staining
Detection of HIV-1 Gag-specific antibodies in immunized mice
-
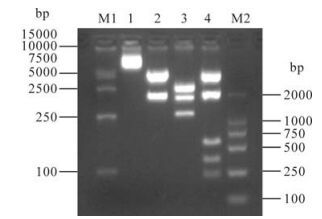
Restriction endoenzyme analysis was used to identify the correct insertion of the gag gene into the pcDNA3.1 (+) vector. 5.4 kb and 1.5 kb of fragments were obtained when pcDNA3.1 (+)-gag was digested with KpnⅠ/XhoⅠ. 4.3 kb, 1.6 kb, 0.5 kb, 0.3 kb and 0.2 kb of fragments were obtained after digestion with Pst Ⅰ. KpnⅠ/SalⅠdigestion gave 2.3 kb, 2.2 kb, 1.5 kb and 0.9kb bands (Fig. 1). The results were consistent with expectations and indicated that the gag gene was correctly inserted into the pcDNA3.1 (+) vector.
-
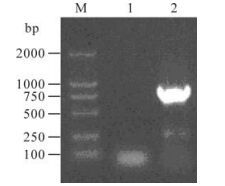
RT-PCR was preformed using the RNA from LLC-MK2 cells infected with rSeV-control and rSeV-gag. A specific fragment of 0.8 kb was ampli-fied in LLC-MK2 cells infected with rSeV-gag while no band was found in the rSeV-control infected cells (Fig. 2). It indicated that gag gene was inserted into genome of rSeV vector.
-
Expression of the Gag Protein in DNA and rSeV vaccines was analyzed by Western blotting. The modified gag gene in plasmid pcDNA3.1(+)-gag was expressed correctly and efficiently after transfecting to LLC-MK2 cells. As shown in Fig. 3, a specific band of 55 kDa could be detected in pcDNA3.1 (+)-gag transfected cells. The recombinant virus rSeV-gag gave a similar level of expression of Gag protein, while no bands could be seen in mock cells.
-
To find a better immunization scheme for these two vaccines, immunogenicty with single or combined modality was compared. As shown in Fig. 4, two individual immunizations with the DNA or rSeV-gag vaccine alone could induce low levels of Gag-specific CTL responses, while DNA vaccine priming and rSeV-gag boosting elicited high frequency of and long-lasting CTL responses targeting Gag. One week post immunization, the percentage of IFN-γ secreting CD8+ T cells in total CD8+ T cells reached 13.1% ±2.2% in the combined immunization group. Although the Gag-specific CD8+ T cells level declined with time, 8.1%±5.7% and 2.5±1.3% Gag-specific CD8+ T cells were detected at 5 and 9 weeks post immuni-zation respectively in this group. Only 1.3%±0.3% and 1.9%±0.8% Gag-specific CD8+ T cells could be induced at 1 week post immunization in the DNA group and rSeV-gag group. They decreased to much lower levels at 5 and 9 weeks post immunization in these two groups.
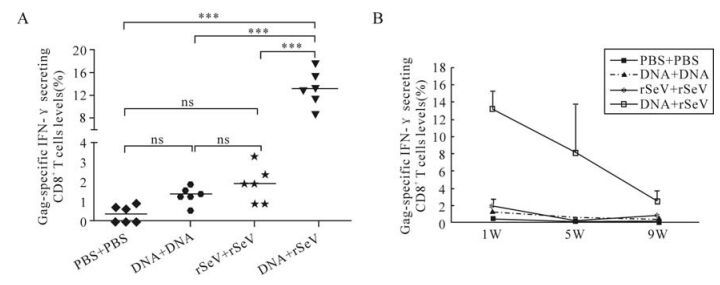
Figure 4. Comparison of Gag-specific CD8+T cells levels in mice immunized with different vaccines. Numbers of Gag-specific CD8+ T cells are shown as percentages of IFN-γ secreting CD8+ T cells in the total CD8+T cells. A: Gag-specific CD8+T cells level in mice at 1 week post immunization. Newman-Keuls Multiple Comparison Test was used for comparison between two groups. ns: P > 0.05; ***: P < 0.001. B: Comparison of Gag-specific CD8+T cells levels in mice at different time point post immunization.
-
Sera of immunized mice were collected at different time points and P24-specific antibodies were detected by ELISA. As shown in Fig. 5, the mice immunized with DNA and rSeV-gag vaccines induced high level of anti-P24 antibodies at 1 week post immunization. The anti-P24 antibodies reached peak levels at 5 weeks post immunization (1: 6 400) and then decreased but still kept high levels compared with other groups. Two Immunizations with DNA or rSeV-gag vaccine did not induce P24-specific antibodies at 1 week post immunization. In mice immunized with DNA vaccine twice, P24-specific antibodies were detected at 5 and9 weeks post immunization, while in mice immunized with rSeV-gag vaccine twice anti-P24 antibodies was undetectable even at 5 and 9 weeks post immunization.

Figure 5. P24-specific antibodies levels in immunized mice. A: Anti-P24 antibodies levels in mice at one week post immunization. Newman-Keuls Multiple Comparison Test was used for comparison between two groups. ns: P > 0.05; ***: P < 0.001. B: Comparison of anti-P24 antibodies levels in mice at different time point post immunization.
Restriction endoenzyme analysis of pcDNA3.1 (+) –gag
gag gene was inserted into rSeV-gag correctly
DNA and rSeV-gag vaccines express HIV-1 Gag protein efficiently
Comparison of gag-specific CTL responses in mice
Comparison of HIV-1 Gag-specific antibodies levels in immunized mice
-
The prevalence of HIV/AIDS poses a severe threat to human health worldwide. Because drugs that could eliminate HIV infection are still not available, an effective vaccine represents the best hope to curtail the HIV epidemic. The objectives of this study are constructing DNA and rSeV vaccines based on prevalent Thailand subtype B HIV-1 gag genes in China and trying to use these vaccines for therapeutic and prophylactic vaccines.
A number of studies have suggested that plasmid DNA is quite effective as a priming or initial immunogen in a bimodal vaccine strategy (2). Expressionof HIV structural proteins in plasmids DNA has been hampered by the fact thattheir expression is dependent on the HIV Rev protein and the Rev-responsiveelement. These proteins could be expressed efficiently only in the presence of rev gene in the plasmid DNA, otherwise their expression level is very low. Changes in the codon usage of HIV structural proteins to those employed by highly expressed human codons resulted in increased Rev-independent expression (3, 14, 25). In order to increase the expression level of Gag in DNA vaccine, the codons of the consensus Thailand B gag sequence from Henan were modified according to mammalian codon usage. And it was verified that expression level of gag was improved largely by codon-modification (data not shown). The codon modified consensus gag gene was used to construct the DNA vaccine.
The SeV vector emerged as a member of a new class of viral vectors with the development of reverse genetics technology. SeV is an enveloped virus with a negative-sense RNA genome. It causes fatal pneumonia in mice, its natural host, but is thought to be nonpathogenic in primates, including humans (11, 21). Some of the extraordinary features of SeV make it particularly suitable for expressing HIV structural proteins. Because its replication cycle is exclusively in cytoplasm, the HIV gene expressed in this vector does not contain a nuclear phase, therefore Rev is not necessary and the gag gene in this vector could be expressed efficiently without codon modification. Because of this, the wild type Thailand B gag gene from Henan province was used to construct a rSev vaccine. It was verified that Gag protein could be expressed well in this vector. The SeV vector used in this study was developed by DNAVEC Corporation. They had confirmed that SIV Gag-expressing rSeV vaccine combined with DNA vaccine could induce high frequency of Gag-specific CTL responses and exerted excellent protective efficacy in macaque models (10, 19). Based on these results we tried to develop vaccines expressing HIV-1 Gag derived from Chinese prevalent strains using the same rSeV vectors. In the studies performed by the DNAVEC Corporation rSeV the vaccines were introduced by intranasal route. In the present study intramuscular immunization was performed for both DNA and rSeV vaccines, and high level of Gag-specific CTL responses and antibodies were elicited in DNA prime/rSeV boost group.
There are many studies in recent years which indicate that the combinations of DNA and recombinant-viral-vector based vaccines are promising AIDS vaccine methods because of their potential for inducing cellular immune responses (2, 9). However, in late 2007 the failure of Merck rAd5 Phase Ⅱ proof-of-concept STEP study began to influence thinking on the design of T-cell vaccines. The candidate may have failed for several reasons. It may not have contained the right mix of HIV components. Alternatively, the vector (Adenovirus type 5) may not be robust enough. Scientists at Merck are looking hard at these issues and collaborating with others to help decipher the data. Given that T cells do not work until infection has occurred, prevention of infection is very likely too high a bar for a T cell vaccine. The more realistic goal for such a vaccine would be to significantly reduce the viral load of individuals who have been infected with HIV. So in our design the vaccines would be first used for therapeutic immunization to control HIV repli-cation in the future clinical trials. If this works then prophylactic immunization could be studied further.
In the present study we compared the immuno-genicity of DNA and rSeV-gag vaccines applied in single or combined vaccines. Our results demonstrated that combined vaccines in the DNA prime/rSeV-gag boost vaccination regimen induced the strongest and most long-lasting Gag-specific CTL and antibody responses compared with single vaccines. The CTL responses were induced earlier compared with humoral immune responses. It reached a peak at one week post immunization and then decreased with time. Even at 9 weeks post immunization comparative Gag-specific CD8+T cells could be detected in this group. The Gag-specific antibody peak occurred at 5 weeks post immunization in this group. DNA vaccine or rSeV-gag vaccine alone elicited a low frequency of HIV-specific CTL responses. Gag-specific antibodies were not detected in mice immunized with rSeV-gag vaccine alone, while in mice immunized with the DNA vaccine antibody responses could be induced and the antibody peak occurred at 9 weeks post immuniztion. As we expected the DNA vaccine showed low immunogenicity when used alone, but why when the rSeV vaccine was used alone could it not induce a high level of humoral response? We speculate that anti-SeV antibodies were elicited after the first immunization. These antibodies would interfere with the effect of the second immunization. We performed another test to verify this hypothesis. Mice were immunized with 1.8 ×107 CIU rSeV vaccine, 3 weeks later anti-SeV antibodies were detected, the titer was 1: 919. This indicated that though not high, anti-rSeV antibodies were induced and may disturb the further use of this vaccine. Another possible reason is the antigen presentation pathway of SeV is different from other vectors; further studies are needed to demonstrate the mechanism.
In conclusion our data demonstrated that DNA prime/rSeV-gag boost vaccination regimen could induce high level of cellular and humoral immune responses in mice.














 DownLoad:
DownLoad: