-
Over the last two decades, almost all the new emerging infectious diseases have involved zoonotic or species-jumping by the infectious agents. RNA viruses with high adaptation ability to environmental changes are the most prominent emerging pathogens such as the highly pathogenic H5N1, Nipah virus, Hendra virus, Ebola virus.
Paramyxovirus Tianjin strain with high hemaggluti-nation titer was isolated from the lungs of common cotton-eared marmoset that died during an outbreak of severe respiratory infection in the marmoset colonies in the Experimental Animal Center in June, 1999 (5). Electron microscopic observations, serodetection and hemagglutinin-neuraminidase (HN) nucleotide sequence analysis indicated that the virus belonged to the Respirovirus genus in the Paramyxovirinae, the family Paramyxoviridae and has a close relationship to Sendai virus (8). Serodetection indicated that the majority of the personnel working in the Center have detectable antibodies against this virus, even people (14/40) who had never come in contact with mar-mosets. We also detected the antibodies in sera of young children with acute respiratory tract infection. The IgM positive rate was 19.28%. These results suggested that paramyxovirus Tianjin strain may have a close relationship with humans, and might be a common respiratory virus that infects both human and marmoset.
The complete genome sequence of paramyxovirus Tianjin strain has now been determined (GenBank accession number EF679198). The genome is 15384 nt in length and encodes at least six structural proteins including two membrane glycoproteins: hemag-glutinin-neuraminidase (HN) and fusion protein (F), which are responsible for viral attachment, penetration and release; three nucleocapsid-related proteins inclu-ding nucleocapsid protein (NP), phosphoprotein (P) and large protein (L), which cover the negative single strand genomic RNA and are responsible for RNA transcription and replication; and matrix protein (M), which is an internal membrane protein, likely me-diating packaging of the nucleocapsid into the viral envelope during virion assembly.
Although phylogenetic analysis of the nucleotide sequence of HN and F gene indicates paramyxovirus Tianjin strain has closer relationship to Sendai virus BB1 strain than the other Sendai viruses, it locates in a unique evolutionary branch (4, 9). It also exhibits the divergent pathogenicity from the classic Sendai viruses in virulence and host range. The common Sendai viruses usually cause lethal pneumonia in mices. However, experimental mice and rats bred in the same Animal Center with the common cotton-eared marmosets had never suffered from the respi-ratory disease in 1999. It is rarely reported that Sendai virus causes the lethal lower respiratory tract infection in marmoset colony (3). Therefore, we theorize that the paramyxovirus Tianjin strain might be a new variant of Sendai virus with high pathogenicity to primates but low pathogenicity to rodents. To date, although the factors that control the host tropism and pathogenesis of Tianjin strain have not yet been elucidated it is certain that HN protein plays a key role. To further understand the relationship between HN structure and function of paramyxovirus Tianjin strain, three segments of the extracellular domain of HN protein: HN1 (61aa-260aa), HN2 (253aa-452aa) and HN3 (375aa-575aa) were expressed. Their antigeni-city, hemaglutination activity and the reactivity to the standard antisera against influenza virus type A and type B were analyzed.
HTML
-
Paramyxovirus Tianjin strain was first isolated from the lungs of a common cotton-eared marmoset that died from severe lower respiratory tract infection in 1999. The virus was passaged in 9-11 days em-bryonated chicken eggs and the allantoic fluids with hemaglutination titer 1:320~1:1280 were collected. The fluids (350mL) were clarified by centrifugation at 1 000 ×g (Beckman Avanti J-25, USA) for 30 min at 4℃. Supernatants were then centrifuged for 3 h at 199 500 ×g (Beckman Optima LE-80k, USA) at 4℃. The pellets were resuspended in a total of 20 mL of PBS. The viral solution was divided into aliquots and stored-70℃.
-
The viral genomic RNA was extracted from purified virus by Viral RNA kit (OMEGA bio-tek, USA) and three 600 bp segments: HN1, HN2 and HN3 covering the extracellular domain of the HN gene were amplified by the reverse transcription PCR (RT-PCR). Three pairs of oligonucleotide primers (Table 1) were designed based on the genome of paramyxovirus Tianjin strain in GenBank. Amplified target DNA segments were ligated into pET28a vector between EcoR Ⅰ and Sal Ⅰ sites and the expression plasmids pET28a-HN1, pET28a-HN2 and pET28a-HN3 were constructed. Subsequently, E.coli Top10 were trans-formed by the recombinant plasmids.

Table 1. Primers for RT-PCR amplification
-
After identification by PCR and double restriction digestion (EcoR Ⅰ and Sal Ⅰ), E. coli BL21 (DE3) were transformed by the plasmids pET28a-HN1, pET28a-HN2 and pET28a-HN3. Transformed bacteria were cultivated in Luria broth (LB) with kanamycin at a final concentration of 100µg/mL at 37℃until induction. Expression of recombinant proteins HN1 (rHN1), HN2 (rHN2) and HN3 (rHN3) with N-terminal 6×His tag was induced for 3 h by addition of final concen-tration of 1 mmol/L isopropylthiogalactopyranoside (IPTG) and rHNs were purified with Ni-NTA column (Promega) according to the manufacturer's manual. The recombinant proteins were quantified by Broadford assay and analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
-
PcAb against paramyxovirus Tianjin strain was produced routinely in Japanese White Rabbits by immunization with purified virus. Anti rHN1, rHN2 and rHN3 mouse polyclonal antisera were prepared in specific pathogen free (SPF) BALB/c mice by immunization with the purified rHN1, rHN2 and rHN3.
-
In dot blots, 5 μg of each rHNs were bound to polyvinylidene difluoride (PVDF) membranes. In western blots, each of the three solutions individually containing the same amounts of purified rHN1, rHN2 or rHN3 was mixed with same volume of 2×SDS loading buffer, and boiled for 5 min. After electrophoresis on a 12% SDS-PAGE, rHNs were transferred to PVDF membranes. The lane of protein marker was separated and stained by amino black solution. The membranes were immersed in PBS-5% nonfat milk at 4℃ overnight. The membranes were incubated with specific rabbit PcAb (1:500 dilution) for 1 h at room temperature, then washed three times with PBS-0.1% Tween-20 for 10 min each time then incubated with peroxidase-conjugated goat anti-rabbit IgG (1:1000 dilution) and the protein bands were visualized by 3, 3-diaminobenzidine (DAB) color reagents.
-
Hemaglutination (HA) test and hemaglutination inhibition (HI) tests were utilized for detecting hemaglutination of rHNs and hemaglutination inhibition by their antisera.
The HA test was carried out as follow. Four solutions containing 40mg/L rHN1, rHN2, rHN3 and bovine serum albumin (BSA, as the negative control) were serially diluted (1:2 ratio) with the elution buffer (100 mmol/L HEPES, 500mmol/L imidazole, 6 mol/L urea, pH7.5), and incubated with 1.0% chicken RBC in a multi-well plate for 1h at room temperature. The hemagglutination titers of rHN1, rHN2 and rHN3 were determined as the highest dilution of each solution that can cause an obvious "++" (50%) hema-glutination.
HI test was described briefly as follow. The 1:2 serial dilutions of the antisera were mixed with four hemagglutination units of paramyxovirus Tianjin strain, and then incubated with 1.0% chicken RBCs at room temperature for 1 h. The hemagglutination inhibition titer was determined as the highest dilution of serum that completely inhibited hemagglutination.
-
Serological reactivity of standard antisera against influenza virus type A (A/NEW CALEDONIA/20/99 (H1N1), A/WYOMING/03/2003(H3N2)), influenza virus type B (B/Hong Kong/1434/2003, B/Jiangsu/10/ 2003) and three rHNs were measured by enzyme linked immunosorbent assay (ELISA). 96-well plates were coated overnight at 4℃ with rHN1, rHN2 or rHN3 at a concentration of 5.0 µg/mL in coating solution (Na2CO3 1.59g, NaHCO3 2.93g in 1.0 L of distilled water, 6 mol/L urea, pH9.6). Antigen-coated plates were washed three times with PBS (pH 7.4) and blocked with 3% BSA at 37℃ 1.5 h. After washing three times with PBS, plates were stored at 4℃ for later use. A 1:4 dilutions of the standard antisera were incubated in a humidified chamber for 30 min at 42℃, and the plates were then washed three times with PBS containing 0.1% Tween 20. HRP-conjugated goat anti-human IgG was added. After incubation at 42℃ for 30 min, the plates were again washed three times with PBS containing 0.1% Tween 20. A substrate of 3, 3', 5, 5'-tetramethylbenzidine (TMB)/H2O2 was added and color developed for about 20 min.
Propagation and purification of the virus
Viral RNA extraction and construction of trun-cated HN expression plasmids
Recombinant protein expression and purification
Preparation of polyclonal antibody (PcAb)
Antigenicity identification of rHNs
rHNs bioactivity detection
Serological reactivity
-
HN1, HN2 and HN3 segments covering HN gene extracellular region of paramyxovirus Tianjin strain were amplified by RT-PCR. As expected, three 600bp segments were obtained (Fig. 1). The PCR products were cloned into pET28a vector and the recombinant plasmids pET28a-HN1, pET28a-HN2 and pET28a-HN3 were identified by double restriction digestion (EcoR Ⅰ and Sal Ⅰ). (Fig. 2 A and B)
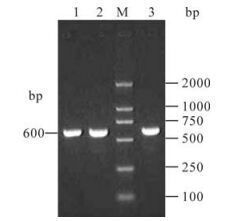
Figure 1. RT-PCR amplification of HN1, HN2, HN3 segments from cDNA of paramyxovirus Tianjin strain. Lane 1, HN1; Lane 2, HN2; M, DNA Marker DL2000; Lane 3, HN3.
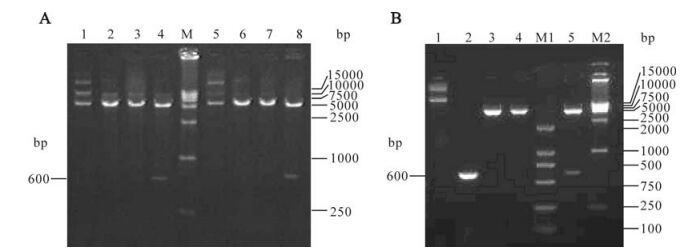
Figure 2. Restriction endonuclease analysis of recombinant plasmid pET28a-HN1, pET28a-HN2, pET28a-HN3. A: Lane 1, Recombinant plasmid pET28a-HN1; Lane 2, EcoR Ⅰ digestion of pET28a-HN1; Lane 3, Sal Ⅰ digestion of pET28a-HN1; Lane 4, EcoR Ⅰ and SalⅠ digestion of pET28a-HN1; M: DNA Marker DL15000; Lane 5, Recombinant plasmid pET28a-HN2; Lane 6, EcoR Ⅰ digestion of pET28a-HN2; Lane 7, SalⅠ digestion of pET28a-HN2; Lane 8, EcoR Ⅰ and Sal Ⅰ digestion of pET28a-HN2. B: Lane 1, Recombinant plasmid pET28a-HN3; Lane 2, PCR amplification of HN3; Lane 3, EcoR Ⅰ digestion of pET28a-HN3; Lane 4, Sal Ⅰ digestion of pET28a-HN3; Lane 5, EcoR Ⅰ and Sal Ⅰ digestion of pET28a-HN3; M1 and M2, DNA Marker.
-
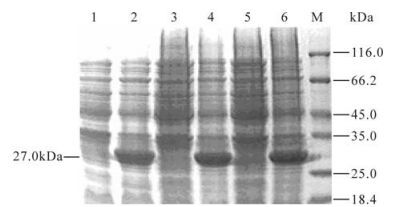
E.coli BL21 (DE) cells were transformed with the recombinant plasmid pET28a-HN1, pET28a-HN2 and pET28a-HN3 and the respective proteins were induced to be expressed highly in inclusion bodies. The proteins were observed as 27 kDa on 12% SDS-PAGE, which is similar to the theoretical molecular weight (Fig. 3). Subsequently rHNs were purified on Ni-NTA columns. ELISA and dot blot analyses showed that rHN1, rHN2 and rHN3 had the similar degree in binding to the specific rabbit anti-Tianjin strain PcAb (data not shown). However, the proteins did not bind to the PcAb to the same degree in Western blots. The antigenicity of rHN2 is the strongest as measured by the intensity of the color of bands and rHN3 had a lower intensity while rHN1 did not produce a visible reactive band (Fig. 4).
-
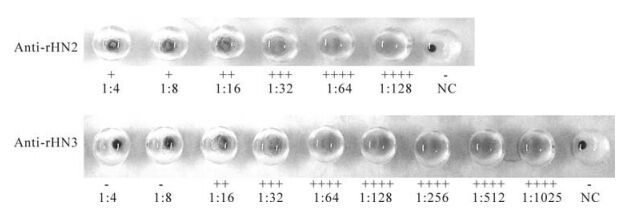
Hemaglutination assays were conducted on rHN1, rHN2 and rHN3. The protein rHN3 showed the higher hemaglutination titer of 1:48 than rHN2 (1:14) while rHN1 exhibited a poor titer of 1:6 (Fig. 5). Hemaglutination inhibition titers of anti-rHN2 and anti-rHN3 were < 1:4 and 1:8 respectively (Fig. 6). Anti-rHN1 could not inhibit viral hemaglutination.
-
To assess if rHN1, rHN2 and rHN3 could be used as specific antigen for the detection of virus in clinical specimens, ELISA was employed to test the cross reactivity between the standard sera against type A and type B influenza virus and the three rHN proteins. Both the standard sera showed positive cross reactions to rHN1, rHN2 and rHN3 (data not shown).
Amplification of HN gene and identification of recombinant expression plasmids
Expression of recombinant proteins and identifi-cation of their antigenicity
Hemaglutination of rHNs and hemaglutination inhibition of mouse rHNs antisera
Cross reaction between standard antisera and the three rHNs proteins
-
The hemagglutinin-neuraminidase (HN) of Sendai virus, a type Ⅱ transmembrane glycoprotein on the surface of the virus particle is a tetrameric spike consisting of an internal membrane domain (aa1-aa35), a transmembrane region (aa36-aa60) and the ectodo-main (aa61-aa575). The HN protein has three discrete activities: the sialic acid-containing receptor binding mediating hemagglutination (HA), neuraminidase (NA) and fusion.
Paramyxovirus Tianjin strain was isolated from the lungs of common cotton-eared marmoset that died from the severe respiratory infection in June 1999 and was a novel strain of Sendai virus with a high pathogenicity to primates and a low pathogenicity to rodents. To partly elucidate the relationship between the HN structure and function of the Tianjin strain, three segments of the ectodomain HN protein: HN1 (61aa-260aa), HN2 (253aa-452aa) and HN3 (375aa-575aa) were expressed and their antigenicity and hemaglutination activities were determined.
By ELISA and dot blots, rHN1, rHN2 and rHN3 showed similar binding properties to anti-virus PcAb. However, in Western blots there apparently the proteins were bound differentially to the PcAb. The levels of antigenicity for the three proteins appears to be in the order of rHN2 > rHN3 > rHN1. In ELISA and dot blot tests, rHNs binding to the solid phase such as polyvinylidene difluoride (PVDF) membrane or polystyrene were likely to have been partially renatured when the denaturing agent was removed during washing and blocking of the membrane (plate). Therefore, the rHN proteins may have been recog-nized by the conformation-specific antibodies and by the linear epitope-specific antibodies in the PcAb. However, the rHN proteins were completely dena-tured by boiling prior to analysis by Western blots and may have bound to only the linear epitopes-specific antibodies. The intensity of color in Western blots depends on the amount of immunodominant linear epitope (11, 12). Our study indicated that amino acid residue 253 to 452 (HN2) contains more linear epitopes exposed on the surface of the virion than segment HN1 and HN3. The denatured rHN1 could not bind well to the anti-virus PcAb suggesting that the residue 61 to 260 of native HN protein might not contain linear epitopes or are hidden to inside the HN protein. The amount of linear epitopes in the domain 375aa-575aa (HN3) is somewhere between HN1 and HN2.
The sialic acid-containing receptor binding is one of three main biological functions of Sendai virus HN protein and its activity could be measured by the hemagglutination assays. To date, double receptor binding sites in HN have been found in Sendai virus as well as human parainfluenza virus type 3 (HPIV3) and Newcastle disease virus (NDV) (6, 7, 14). The amino acid residues involved in the formation of two receptor binding sites varies among paramyxoviruses. In Sendai virus, residues 191 and 198 influence the receptor binding activity of site Ⅰ, and residues 520 and 523 are crucial in the formation of site Ⅱ (2). In our study, expressed rHN1 (61aa-260aa) and rHN3 (375aa-575aa) contain key residues which form the receptor binding site Ⅰ and site Ⅱ, respectively. The lower hemagglutination titer of rHN1 (1:6) under denatured condition than that of rHN3 (1:48) suggested the receptor binding activity of site Ⅰ may be more dependent on three-dimensional conformation of HN protein than that of site Ⅱ. The hemaggluti-nation titer 1:14 shown in rHN2 indicated the domain (253aa-452aa) is also associated with the receptor binding activity of HN.
The practice so far is to apply either the complete inactivated virus or viral lysates as antigen in the majority of the serological tests to detect Sendai virus (1, 10, 13). Extensive cross reactions exist between Sendai virus and other paramyxoviruses. We have attempted to find more specific antigens than the classic ones from rHN1, rHN2 or rHN3. Indirect ELISA tests were performed to exam the cross reactivity between the standard anti influenza virus type A, B antisera and the three rHN proteins. Positive cross reactions indicated that rHN1, rHN2 and rHN3 are unsuitable for specific serodetection. Recently, rHN proteins have been used to prepare McAbs that can specifically recognize HN of paramyxovirus Tianjin strain or Sendai virus. They might be helpful to set up the more specific serological detection methods to investigate the epidemic of paramyxovirus Tianjin strain in human.















 DownLoad:
DownLoad: