-
The mumps virus (MuV) causes a systemic illness spread by virus-containing droplets from the upper respiratory tract. Infection with mumps is asympto-matic in one-third of cases. The disease is generally self-limiting and usually characterized by parotitis and mild nonspecific symptoms, but it has the capacity to invade the visceral organs and central nervous system (CNS).
The mumps virus is a member of the Rubulavirus genus of the family Paramyxoviridae. The virus contains a negative-sense RNA genome of 15 384 nucleotides that consists of seven transcription units. The gene order is as follows: nucleoprotein (N), phosphoprotein (P), and matrix (M), fusion (F), small hydrophobic (SH), hemagglutinin-neuraminidase (HN), and large (L) proteins. The SH gene (57 amino acids) is the most variable segment of the MuV genome, with a missense to silent mutation ratio of 2.0 (cf. a ratio of <0.5 for other MuV genes). Phylogenetic analyses are, therefore, mainly based on the SH gene sequence. Sequence comparison based on the SH gene has led to the definition of distinct MuV genotypes (6, 11). As of 2007, 13 genotypes known as A-M had been described (12).
In China, mumps outbreaks due to the genotype F virus still occur (8, 17). Infection with genotype F mainly results in parotitis and rarely in meningitis. No isolates belonging to genotype F were found in cere-brospinal fluid (CSF) samples. Even though a definitive study cannot be performed due to the lack of neuro-pathogenic isolates in China, we speculate that geno-type F (including SP strain) is likely to cause a milder illness than meningitis. However, this genotype may exhibit neurovirulence in some cases. In this study, we determined the entire nucleotide sequence of a genotype F wild-type mumps virus isolated from a patient in ShiPing, China and compared it with those of previously reported strains.
HTML
-
Five throat swab samples were collected from patients with parotitis in ShiPing, China in March, 2006. Vero cells were cultured in five tissue culture flasks in 25cm2 of minimum essential medium (MEM) supplemented with 5% fetal calf serum (FCS). A 1 mL throat swab sample was inoculated into Vero cells. After contact for 1 hr at 37 ℃, samples were removed, and MEM supplemented with 2% FCS was added. Within three passages, samples that induced mumps virus-specific cytopathic effects (CPEs) were consi-dered to be positive for virus isolation. Samples that did not induce CPEs through three passages in Vero cells were considered to be negative. Supernatants from the inoculated wells showing CPEs were harvested. The supernatant was identified by a culture-neutralization test using the Institute of Medical Biology, Chinese Academy of Medical Sciences and Peking Union Medi-cal College(IMB)mumps virus-antisera. This proce-dure is the standard diagnostic currently used in China.
-
The 36 primers used in RT-PCR are shown in Table 1. Primers were newly designed based on the publi- shed sequence of the UrabeAM9 strain (GenBank ACC# AB000388, genotype B). Many primers were described in the previous reports (4, 6, 7).

Table 1. Primers used for RT-PCR and sequencing of mumps virus genome
RT-PCR, cloning, and sequencing Viral RNA was extracted from culture fluids using a Viral RNA Kit (Qiagen Inc., USA) according to the manufacturer's instructions. The RNA was reverse transcribed into first strand cDNA with the following primer pairs: 1F, 7F, 13F, 19F, 23F, and 29F. These primers covered the entire genome of the mumps virus (Revertaid TM first strand cDNA synthesis Kit). PCR reactions were carried out using Taq polymerase (TakaRa Dalian Co., Japan) and other essential reagents. The cycling condi-tions consisted of an initial denaturation at 94 ℃ for 5 min, 35 cycles of 94 ℃ for 30 s, 54 ℃ for 30 s, and 72 ℃ for 2 min, and a final extension step for 10 min at 72 ℃ in a thermal cycler. These amplified products were gel-purified and then cloned into a PMD18-T vector (TakaRa Dalian Co., Japan) according to standard procedures. Recombinant plasmids confirmed by PCR were sequenced by Sangon Corporation.
-
Sequencing results analyzed and compared using the MEGA4.1 (Rainbow Technologies, INC) software package to define the genetic variations and relation-ships with other strains were obtained from GenBank. The complete sequence of the SP strain was submitted to the GenBank under accession number GenBank ACC#DQ-649478, genotype F.
The strain/isolate names and GenBank accession numbers for sequences used in this work are as follows: Complete genome sequences: 87-1004 (AF-314560), 87-1005 (AF314562), 88-1961 (AF467767), Biken (AF314561), Dg1062/Korea/98 (Korea98) (AY 309060), Drag94 (AY669145), Glouc1/UK96 (UK96) (AF280799), JERYL LYNN-2 (vaccine strain, JL2) (A F345290), JERYL LYNN-5 (vaccine strain, JL5) (AF-338106), L-Zagreb vaccine (vaccine strain) (AY685920), Miyahara (AB040874), PetroNov (AY681495); SP (DQ649478); Smith-Kline Beecham (Smith, vaccine strain) (AF314559), SIPAR 02 (AF314558), L3/Russia/ Vector, (AY508995).
SH gene sequences (SA702Ja99, AB056145), Ya-maguchi99 (Ab105482), TokyoM-21 (AB105478), Mu-VS-ISR05-98649-G5 (AM293334), UK(AF280799); New Jersey (US2006), L-Zagreb (AY685920), L3/ Russia (AY508995), Islip1-UK97(AF142766), Kent1-3UK97 (AF142767), Se50647 (U50284), SEVII (U 50 291), DK8307 (AF365919), DK8206 (AF365897), DK8306(AF365918), Fukuoka49-JPN.00 (AB105483), TokyoS-III-10-JPN.01 (AB105480), TokyoM-50-JPN. 00 (DQ136174), 36-1079 (AB116011), Minsk.Be-larus/ 10.02 (DQ136175), Minsk.Belarus 44.01 (DQ 136174), MinskBelarus09.03, (DQ25004), MuVs-PAL-04-86994 (AM293337), 871004 (AF314560), 871005 (AF314-562), 88-1961 (AF467767), Biken (AF314561), Kor98 (AY309060), DRAG94 (AY669145), Glouc1/ UK96 (UK96) (AF280799), JL2 (F345290), JL5 (AF 338106), PetroNov (AY681495), SP (DQ649478), Yeo ju1503 (AY048995), 776274 (AF526410), SA475Ja 97 (AB0 56148), AS97-8 (AF180376), IS98-58 (AF 180384), SIPAR02 (AF314558), MUVs-PAL04-89033 (AM29 3338), Edingburgh2 (X63711), Edingburgh6 (X637 12), Edingburgh4 (X93177), Belfast (X63709), Bri-stoll (X63713), Polos-Portugal-96 (Y08214), Wlz1 (Z77158), Wsh1 (Z77160), Wsh2 (z81005), Wsh3 (U 80435), Wlz2 (Z77161), Wlz3 (Z77159), Zhejiang 06-30-10 (EF102880), UK99-102x13 (AY380065), UK99-190 (AY380068), MuVs-ISR04-94507 (AM 293339), 1700 (AB115996), MP94-H (AB003417), TK087 Ja 97 (AB05614).
Virus isolation
Primers
Sequence analysis
-
One virus strain known as SP was isolated with CPE characteristics in Vero cells. This strain was confi-rmed by a culture-neutralization test and RT-PCR.
-
Two mumps virus phylogenetic trees based on the 316 nucleotide region of the SH gene and the com-plete genome were generated by the neighbor-joining method. In Fig 1, the SP strain was closely related with Wlz2 and Wsh3 (Fig. 1). The data showed that these strains may belong to the same genotype (geno-type F). In Fig. 2, the no closest genetic relationship to SP strain is compared with the complete genome of JERYL LYNN-2 and JERYLLYNN-5.
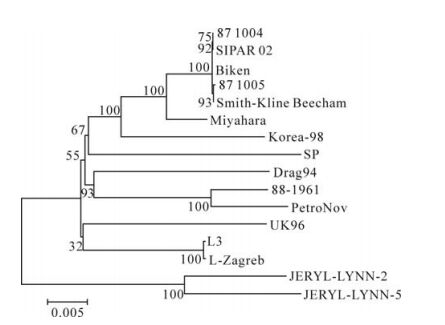
Figure 1. Mumps virus phylogenetic tree generated by the neighbor-joining method, based on the complete genome of mumps virus. The phylogenetic tree was constructed by the neighbor-joining method with MEGA version 4.1 software package, and the reliabilities indicated at the branch nodes were evaluated using 500 bootstrap replications. The length the horizontal branches reflects the phylogenetic distance.
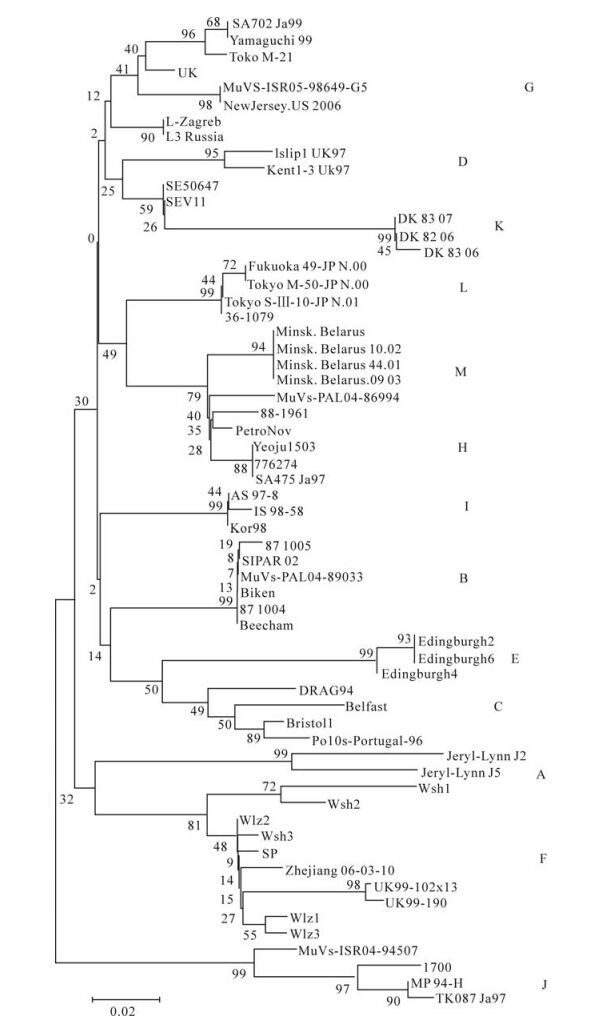
Figure 2. Mumps virus phylogenetic tree generated by the neighbor-joining method, based on a 316 nucleotide region of the SH gene. The phylogenetic tree was constructed by the neighbor-joining method with the MEGA version 4.1 software package and the reliabilities indicated at the branch nodes were evaluated using bootstrap 500 replications. The length the horizontal of the branches reflects phylogenetic distance.
-
The complete sequence of the SP strain was sub-mitted to the GenBank under accession number Gen-Bank ACC#DQ-649478, genotype F. The genome of the SP isolate was 15 384 base pairs in length. Similar to the other 15 strains, it is organized around eight major proteins. The homology with the other 15 mumps virus strains was 96%-93.2%. Amino acid variations were observed.
-
The homologies of the SP strain to 15 other mumps virus strains in the region of the N and L genes were 94.2% to 95.8% and 94.1% to 96.6%, respectively (1 650 nt); the amino acid sequences were 97.2% to 98.9% and 97.8 to 99.2% homologous to the 15 other strains, respectively. For the comparison of the P protein, two extra guanosine residues were inserted at a G-rich region as shown in other reports (3). Faithful transcription without the insertion of a P gene yields the V protein, which may be important for the viral multiplication of the mumps virus in cells. The P and V genes were 92.3% to 96.2% and 92.7% to 96.7% homologous to the 15 other strains, respectively, at the nucleotide sequence levels; the amino acid sequences were 95.4% to 98.3% and 93.3% to 97.3% homo-logous to the 15 other strains, respectively. These data show that the amino acid sequences were mostly conserved, particularly at the NP and L genes (Table. 2), and they confirm findings from a previous report (2).

Table 2. Nucleotide and amino acid homology comparison between SP and other Mumps virus isolates
The N protein also plays a role during viral assembly through interactions with the matrix protein. Antigenic sites of the MuV N protein have been found to reside within its C-terminus in amino acid regions 412-475 and 475-549 (5). We found that the deduced sequence of the SP strain was quite different when compared with vaccine strains belonging to genotypes A or B. Instead, it was more homologous to the L-Zagreb strain (Fig. 3).
-
The SP strain was 93.7% to 96.1% and 92.2% to 96.6% homologous to the 15 other mumps virus strains at the regions of the F and HN genes, respec-tively. The amino acid sequences were 95.2% to 97.4% and 97.2 to 99.1% homologous, respectively. The amino acid sequences of the HN gene were particularly similar to the JL5 and JL2 strains (vaccine strain, A genotype), with 95.4% and 96.2% homology, respectively. The M gene was 94.7% to 97.1% homologous to the 15 other strains at the nucleotide sequence levels, whereas the amino acid sequence was 98.9% to 99.7% homologous to the 15 other strains. Our data thus show that the M gene was mostly conserved (Table. 2).
So far, known epitopes neutralizing antibodies against the MuV are located in the HN protein in amino acid regions 265-288, 329-340, and 352-360. The HN protein, which is important for receptor binding, neuraminidase activity, and promotion of fusion protein function is isolated at the virion surface and provides the major target for the humoral immune response against MuV infection (5). In the deduced HN protein sequence of the SP strain, the antigenic epitopes were quite different than those from vaccine strains belonging to genotypes A or B. Instead, they were identical to those from the L-Zagreb strain (Fig. 4).
A genotype-specific motif at positions 28-30 in the SH gene was observed in the form of Ile-Met-Leu.No similar patterns were observed in other genotypes (10, 14, 15).
Virus isolation and identification
Phylogenetic analysis
Characteristics of the entire nucleotide sequence
Nucleocapsid–associated protein (N, P, V, and L genes)
Membrane-associated protein (M, F, and HN)
-
Live attenuated mumps vaccines have been in use since the late 1960s. Eleven mumps vaccine strains (Jeryl Lynn, Urabe, Leningrad-3, L-Zagreb, Rubini, Hoshino, Miyahara, Torii, NKM-46, S-12, S79, and RIT 4385) have been used throughout the world, although some are employed only on a limited scale. These strains belong to just three clusters: JL 2, JL 5, S79, and Rubini belong to genotype A; Hoshino, Miyahara, and Urabe AM9 belong to genotype B; L-Zagreb belongs to genotype D. The entire genomic sequence was determined for only some of these strains (e.g., Jeryl Lynn, L-Zagreb, and Miyahara). All vaccine preparations utilizing the same parent strain are not identical due to differences in passage history, cell substrates, and manufacturing processes. Outbreaks have not been completely eliminated, even in vaccinated populations. Mumps virus variants may differ in biological characteristics like antigenic reactivity. Therefore, investigation of the genetic diversity of isolates is important (13). However, a limited number of full-length nucleotide sequences of mumps viruses have been determined, and insufficient information regarding biological function is available. Because of this problem, whole genomic sequencing of the isolated SP strain was performed using RT-PCR to demonstrate that the M, L, and NP genes were mostly conserved. The SH gene was the most variable. When compared to the vaccine strains, antigenic variability was observed in the SP strain.
The homologies of the HN and N proteins of the SP strain to 15 other mumps virus strains ranged from 97.2 to 99.1% and 97.2% to 98.9%, respectively. Relatively high divergence was still observed with respect to JL5 and JL2 (4.6% and 3.8%), and anti-genic epitopes or sites were quite different than those from vaccine strains belonging to genotypes A or B. Because of the high mutation rate of viral RNA-dependent RNA polymerase as well as their quasis-pecies population nature, all RNA viruses show considerable genetic plasticity. The MuV has a nuc- leotide substitution rate in the range of 103 to 104 sub-substitutions/site /per year, which is typical for RNA viruses. High rates of molecular evolutionary change do not necessarily translate into high level of antigenic variation. It is generally believed that the MuV is serologically monotypic, although cases of infection of vaccinated individuals have been reported. These cases are typically attributed to vaccine failures, but some laboratory data as well as cases of rein- fection suggest the existence and significance of virus strain or genotype-specific neutralizing antibodies (5).
In the present study, we raise the question of whether a vaccine containing one genotype of the mumps virus is optimal for protection against other genotypes of the mumps virus. The MuV can be divided into 13 genotypes, but the MuV vaccine only contains a limited genotype. The S79 strain (with HN and N sequences identical to those of JL2, data not shown) has been used in China for vaccination for more than 20 years, and the F genotype mainly circu-lates in China. This vaccine may therefore be insuf-ficient for effective protection against the F genotype of the MuV. Further studies will be necessary to clarify this issue.
The amino acid sequence of the HN gene at posi-tion 335 is known to be related to the interaction with host cells. Previous work identified it as a lysine corresponding to the AAA codon, identical to that of other virulent viruses (1). Because the SP isolate was obtained from a child without aseptic meningitis, however, it is likely that this isolate may cause milder symptoms than other neurovirulent strains even though the relative degree of neurovirulence of them could not be determined. Furthermore, its biological and clinical differences remain to be characterized.
The neurovirulence of different mumps viral strains has been related to the cell fusion activity of the viral strain. In particular, viral strains with high neuroviru-lence have been shown to exhibit high levels of cell fusing activity. Neurovirulent viral strains belonging to genotypes A, C, and D have been identified (9), but molecular markers for MuV virulence remain to be determined.
Understanding the nature of the mumps virus is important for controlling the epidemic. To this end, analysis of the complete genome may help us to comprehend the evolution or diversity of the mumps virus. Although extensive studies on the mumps virus have been performed, further study of the relation- ships between biological function, pathogenicity, and genetic information is required.















 DownLoad:
DownLoad: