-
Coronaviruses (CoVs) are large single-stranded positive RNA viruses of the order Nidovirales, family Coronaviridae, genus Coronavirus [4, 32]. They are generally associated with respiratory and enteric infections and have long been recognized as important pathogens of livestock and companion animals[4, 24, 28]. The recent emergence of the severe acute respiratory syndrome coronavirus (SARS-CoV) and an increased awareness of the extent of human coronavirus-associated disease have renewed interest in this group of viruses [10, 22].
Coronaviruses are positive-stranded RNA viruses with genomes ranging in size from 27 to 32 kb. Approximately two-thirds of the Coronaviruses genome encodes the viral nonstructural proteins (Nsp) that are involved in viral RNA synthesis. The majority of these proteins are encoded in two 5'-proximal overlapping open reading frames, ORF1a and ORF1b, translated as polyproteins, pp1a and pp1ab, which are then processed by virus-encoded proteinases into Nsp16 [4, 28, 32, 34]. Many of the Coronavirus Nsps have been shown or are predicted to have enzymatic functions[5, 11, 18, 26]. Nsp16 is predicted to be a S-adenosyl-methionine-dependent 2'-O-methyl transferase which is involved in the formation of viral 5'-cap structures [9, 16]. Whereas the exact role of Nsp16 during CoV replication is still unknown, its functional importance is supported by mutagenesis experiments using a SARS-CoV replicon system. The deletion of the Nsp16 coding sequence blocked RNA synthesis, whereas a single mutation in the catalytic tetrad reduced replicon driven mRNA synthesis to about 10% of the level for the wild type[37].
Mouse hepatitis virus (MHV) is a widely studied model system for Coronavirus replication and pathogenesis. The development of MHV reverse genetics, in particular the method based on the use of vaccinia virus cloning vectors, makes it suitable for analyzing Coronavirus RNA replication and transcription [2, 7, 8, 15, 25, 36]. In this study, we first used reverse genetics to create a MHV-A59 temperature sensitive (ts) mutant Wu"-ts18 (cd), which had the same amino acid change at Nsp16 position 12 (Pro to Ser) as ts mutant Wu"-ts18[29, 33]. Then we cultured the ts mutant Wu"-ts18 (cd) at non-permissive temperatures which "forced" the ts recombinant virus to use second-site mutation to revert from a ts to a non-ts phenotype. Sequence analysis of the revertant provided us genetic information on the functional determinants of Nsp16. This allowed us to build up a more complete model of the functional replication –transcription complex.
HTML
-
Mouse 17 clone 1 fibroblast cells (17Cl-1) were cultured at 37℃ in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL). BHK-MHV-N cells, kindly provided by professor Stuart G. Siddell[8, 31], were cultured in minimal essential medium (MEM) supplemented with HEPES (25 mmol/L), 5% FBS, and antibiotics. Recombinant vaccinia virus (vv inf-MHV-A59) contained a full-length MHV-A59 cDNA (GenBank accession number AY700211)[8]. Wu"-ts18 was a reported ts mutant of wild type MHV-A59[28, 33].
-
In order to obtain the revertants of ts mutants, we first constructed the recombinant ts mutants Wu"-ts18 (cd), which had the same amino acid change as Wu"-ts18. Three nucleotide changes (C20880 to A, C20881 to G, U20882 to C) were made to reduce the likelihood of reversion to the wild-type sequence. Mutagenesis was done using a reverse genetics system described previously[8, 20, 36]. Briefly, two rounds of vaccinia virus-mediated homologous recombination were done using the E.Coli. guanine-phosphoribosy-ltransferase (GPT) gene as a selection marker. Firstly, MHV-Nsp16-GPT-IN recombinant vaccinia virus was obtained by homologous recombination between vv inf-MHV-A59 virus and plasmid pGPT-in-Nsp16. The plasmid pGPT-in-Nsp16 encoded the GPT gene flanked on its left by MHV-A59 nucleotides (nt) 20 317 to 20 817 (up of Nsp16) and on its right by MHV-A59 nt 21 773 to 22 273 (downstream of Nsp16). The Nsp16 protein-coding region within the recombinant vaccinia virus inf-MHV-A59 genome was replaced by the GPT gene using homologous recombination with plasmid pGPT-in-Nsp16. Secondly, the GPT gene within the recombinant vaccinia virus vv inf-MHV-Nsp16-GPT-IN virus genome was replaced by mutated Nsp16 protein-coding regions using homologous recombination with plasmid pGPT-out-Nsp16m, pGPT-out-Nsp16m contains MHV-A59 cDNA sequences corresponding to nt 20 317 to 22 273, including the Nsp16 protein-coding region (ORF2a) between nt 20 846 and 21 742, the codon CCU (nt 20 880-20 882) were changed to AGC by site directed PCR-mutagenesis of plasmid pNsp16. Finally, the recombinant vaccinia virus containing the expected point mutations, vv inf-MHV Wu"-ts18(cd), was obtained after 3 rounds of plaque purification. The identities of plasmids and recombinant vaccinia viruses were confirmed by sequencing the mutated regions.
In vitro transcription from purified, EagI-cleaved DNA of recombinant vaccinia virus vv inf-MHV Wu"-ts18(cd) and the parental virus vv inf-MHV-A59 using bacteriophage T7 RNA polymerase in the presence of an m7G(5')ppp(5')G cap was done as described previously [8, 19, 20, 35]. The full-length MHV genomic RNA was transfected into BHK-MHV-N cells by Lipofectamin 2000 (Invitrogen). At day 1 post-transfection, the transfected BHK-MHV-N cells were mixed with a four-fold excess of fresh 17Cl-1 cells and cultured at 33℃. At day 3 post-transfection, tissue culture supernatants were taken, and continued to assay. Recombinant ts mutant virus Wu"-ts18 (cd) and parental virus inf-MHV-A59 were respectively purified for three rounds at 33℃. Passage 1 virus stocks were obtained by collecting the supernatant of the 17Cl-1 cells infected with the virus from a single plaque. The recombinant ts mutant virus Wu"-ts18(cd) and parental virus in-MHV-A59 were confirmed by sequence analysis.
-
17Cl-1 cells in six-well plates were infected in duplicate at room temperature with 1.0 mL of ten-fold dilutions of plaque purified Wu"-ts18(cd) virus in HBSS containing 50 μg/mL DEAE-dextran and 0.2% BSA. The inoculum was removed after 30 min and the cells were washed with 2 mL of DMEM. Monolayer was overlayed with MEM containing 5% FBS, 100 U/mL of penicillin, 100 μg/mL streptomycin and 0.1% GelriteTM gellan gum (Sigma-Aldrich, Poole, Dorset, UK) and incubated at the appropriate temperature with 7.5% CO2. Infected cells were incubated at 33℃ for 3 days or at 37℃ and 39.5℃ for 2 days. Cells were fixed with 5% formaldehyde in phosphate buffered saline (PBS) and stained with a solution of 0.2% toluidine blue, 0.2% azure blue and 1% boric acid.
-
Recombinant virus were analyzed for replication by using plaque assay as described previously [8, 20, 35]. Briefly, 17Cl-1 cells were infected with viruses at an multiplicity of infection (MOI) of 10 PFU/cell. Following the 30 min adsorption at room temperature, the inoculum was removed, and cells were washed three times with 2 mL of DMEM and supplied with prewarmed 10% FBS-DMEM. Cells were then incubated at 33℃, 37℃ or 39.5℃, virus were collected at different intervals from 1 to 16 h post-infection (p.i.), and virus titers were determined by plaque assay at 33℃. The efficiency of plating (EOP) was determined by dividing the titer at 39.5℃ by the titer at 33℃.
-
Virus stocks were obtained by using virus from a single plaque to infect 17Cl-1 cells to yield passage 1 virus stocks. Viral RNA for RT-PCR and sequencing was obtained by isolating total RNA using the Trizol reagent (Invitrogen) as described by the manufacturer. The entire replicase gene-coding region (ORF1a and ORF1b) of the virus was sequenced using a set of 121 oligonucleotides that were complementary to sequences spaced at approximately 350 nucleotide intervals along the positive and negative-strand copies of the viral RNA (sequences available on request). Five oligonucleotides P17, P31, P46, P61 and P65 were used to amplify the cDNA of the viral RNA with superscript Ⅲ RT (Invitrogen, Paisley, UK). The reaction mixture (20 μL), which contained in addition to pre-supplied buffer, 35 ng of primer, 10-100 ng of infected cell total RNA, 0.5 mmol/L dNTP, 5 mmol/L DTT, 25 U of RNAguard (Amersham Biosciences, Chalfont, UK) and 200 U of reverse transcriptase, was incubated at 55℃ for 60 min and then at 70℃ for 15 min. The cDNA templates were then amplified using eight primer pairs P1/P16, P2/P22, P3/P30, P4/P38, P5/P45, P6/P53, P7/P60 and P8/P64 and thermostable recombinant Taq DNA polymerase. The reaction mixture (100 μL) contained in addition to pre-supplied buffer, 70 ng of primer pair, 1 μL of RT reaction product, 200 μmol/L dNTPs, 2 mmol/L MgCl2 and 2.5 U of DNA polymerase and was incubated at 94℃ for 1 min then 94℃ for 20 s, 50℃ for 20 s, 68℃ for 3 min for a total of 35 cycles and a final extension at 68 ℃. The PCR reaction products were purified by ethanol precipitation using ammonium acetate. Finally, sequence analysis was done using primers P1-P121 in the Invitrogen Company (Invitrogen Beijing, China). Computer-assisted analysis of sequence data was done using the Lasergene bio-computing software (DNASTAR).
Cells and viruses
Construction of recombinant MHV-A59 ts mutant Wu"-ts18(cd)
Plaque assay of recombinant ts mutants Wu"-ts18(cd)
Viral replication analysis
RNA preparation, RT-PCR and sequencing of virus
-
Compared with the wild-type MHV-A59 virus, the ts mutant Wu"-ts18 failed to form plaques or synthesize viral RNA when infection was initiated and maintained at the non-permissive temperature 39.5℃. A single nucleotide change (C20880 to U, ) which induced the amino acid proline (Pro: CCU) change to Serine (Ser: UCU) of Nsp16 at position 12 was identified as the mutation responsible for the ts mutant phenotype. In order to identify the determinants which were important in the function of Nsp16. We first constructed the recombinant ts mutants Wu"-ts18(cd), which have the same amino acid change as Wu"-ts18. Three nucleotides (C20880 to A, C20881 to G, U20882 to C) were mutated to reduce the likelihood of reversion to the wild-type sequence, which "forced" the virus to use second-site mutations to revert from a ts phenotype to a non-ts one.
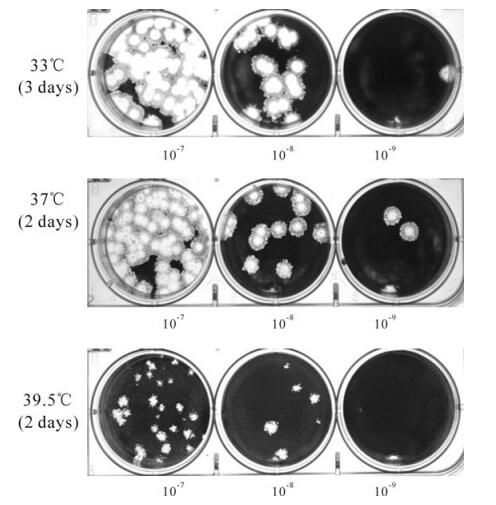
To determine if the constructed mutant viruses Wu"-ts18 (cd) were ts phenotype, the diluted passage one stocks of Wu"-ts18 (cd) which came from one plaque were used to infect 17Cl-1 cells at permissive (33℃ or 37℃) and nonpermissive (39.5℃) temperature. After culturing for 2 or 3 days, the Wu"-ts18 (cd) virus had the same plaque size (6-7mm in diameter) and plaque morphology as the wild-type virus inf-MHV-A59 in 17Cl-1 cells, and it obtained nearly the same high titer of 1-3×109 PFU/mL as the wild-type virus inf-MHV-A59 at the permissive temperature 33℃ and 37℃ (Fig. 1). However, at the non-permissive temperature 39.5℃, compared with the large plaque size formed by inf-MHV-A59, the plaque morphology formed by Wu"-ts18(cd) were heterogenous in plaque size, ranging from almost wild type (6-7mm in diameter), to intermediate (5-6mm in diameter) and small (2-3mm in diameter). The titer of the passage one virus stock was about 1~5×103PFU/ mL, far below that of wild-type virus inf-MHV-A59 (1-5×107PFU/mL).
-
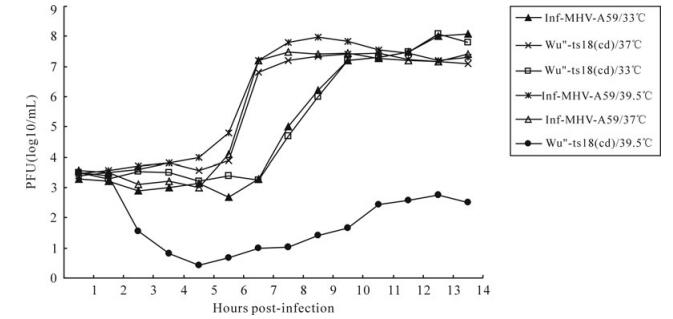
After three rounds of plaque purification, the replication characterization of mutants Wu"-ts18 (cd) at different temperatures were tested on 17Cl-1 cells with an MOI=10. At permissive temperature 33℃ or 37℃, the Wu"-ts18 (cd) mutant was found to be indistinguishable from the inf-MHV-A59 with respect to replication kinetics (Fig. 2); the supernatants from tissue cultures infected with Wu"-ts18 (cd) had reached titers that were equal to or greater than the titers of supernatants from the tissue cultures infected with the parental virus inf-MHV-A59. At 33℃, the Wu"-ts18 (cd) and inf-MHV-A59 were manifested as exponential growth until about 5-6 h p.i., reached the highest titer (~108PFU/mL) at 12 h p.i., which were delayed somewhat compared with results at 37℃, which reached the highest titer (3-5×107PFU/mL) at 7 h p.i.. However, at the non-permissive temperature 39.5℃, inf-MHV-A59 had the normal titer, whereas Wu"-ts18 (cd) had a titer of 3-5×102 PFU/mL.
To analyze the phenotype of Wu"-ts18 (cd) in more detail, the efficiency of plating (EOP) of the two recombinant viruses were calculated by dividing the titer at 39.5℃ by the titer at 33℃ (Table 1). The EOP for inf-MHV-A59 virus was 1.25 which was indistinguishable from the wild type MHV-A59 virus (1.0), confirming that there was no impairment of growth at 39.5℃. In contrast, at 33℃, the recom-binant virus Wu"-ts18(cd) stock had a titer of 3-5×108 PFU/mL and at 39.5℃ had a titer of 1-2×102 PFU/mL, which was an EOP of ~10-7 which was similar to the EOP obtained with the original Wu-ts18 ts mutant. So as expected, the recombinant virus Wu"-ts18 (cd) was indistinguishable from the ts mutant Wu"-ts18 in terms of temperature sensitivity.

Table 1. Temperature sensitivity of recombinant Wu"-ts18(cd)
-
To obtain the revertant of ts mutants Wu"-ts18 (cd), 17C1-1cells were infected with serially diluted Wu"-ts18 (cd) at the non-permissive temperature (39.5℃), 48 h later, revertants were produced, causing large-sized plaques and revertants with noticeably smaller plaques. We picked several plaques into separate wells (including large, intermediate and small size) for plaque purification and after three rounds of plaque purification at the non-permissive temperature (39.5℃), some large (wild-type sized) plaque size revertants consistently formed large wild-type plaques (R1, R3, R4, R7, R8, Fig. 3A); some intermediate and small plaque size revertants reverted to large wild-type plaques (R2, R5, R6), and only one small plaque size revertant consistently formed small plaques (R9, Fig. 3B) which was clearly distinguishable from the wild-type plaques.
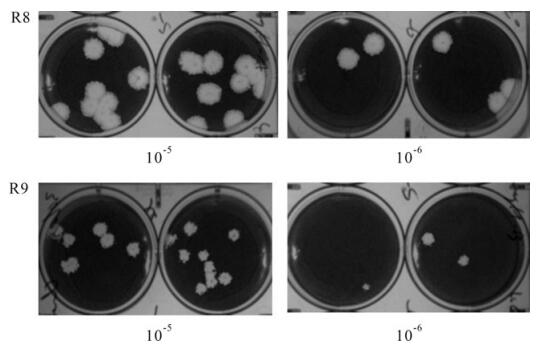
Figure 3. Plaque assay of revertant virus R8 and R9 of Wu"-ts18 (cd) at the non-permissive (39.5℃) temperatures. After 2 days incubated at 39.5℃, R8 plaques were 5-7 mm in diameter, and R9 plaques were 2-3mm in diameter.
After three rounds plaque purification, the revertant plaques were then expanded at 39.5℃ in T25 flasks, and total cellular RNA was obtained. Each isolated virus was subjected to RT-PCR and the entire coding region of the replicase genes (ORF1a and ORF1b) was sequenced as described. Sequencing results (Table 2) showed that all the revertants had retained the introduced Nsp16 position 12 mutation. The most frequent amino acid change that reverted from the ts virus to non-ts phenotype was Nsp16 position 43 (Asn to Ser). Seven of nine revertants had the same amino acid change at Nsp16 position 43 and the presence of this mutation alone reverted from the ts mutant Wu"-ts18 (cd) to non-ts phenotype (such as R1). However, the single amino acid change at Nsp16 position 76 (Lys to Glu) or position 130 (Asp to Asn) produced the same result (revertants R2 and R3). We also obtained revertants with the position 43 mutation plus another mutation in Nsp16, such as R4, R5, R6 and R7. The R8 and R9 had the same mutation at position 43 (Asn to Ser), but they formed large plaque size and small plaque size respectively. Finally, the whole genome of R8 and R9 were sequenced, the results showed there was only one amino acid difference between the revertant R8 and R9. The small plaque isolate R8 revealed an additional mutation in Nsp13 at position 115 (Thr to Ile).

Table 2. Sequence analysis of the Wu"-ts18 (cd) revertants
The functionally uncharacterized Nsp16, has previously been predicted to be a S-adenosyl-Lmethionine (AdoMet)-dependent RNA nucleoside-2'-O-methyltransferase (2'O-MTase) [9, 16, 28, 30]possessing the highly conserved catalytic tetrad (K-D-K-E) that is a hallmark of RNA 2'O-MTases[1, 6, 12, 16]. Structure and sequence comparisons of the 2'O-MTases suggest that the conserved K-D-K-E tetrad formed the active site for the 2'-O methyl transfer reaction. Sequence alignment results showed the amino acid K46-D130-K170-E203 of MHV-A59 Nsp16 corresponded to the conserved K-D-K-E active site. It was notable that the most mutations of the revertants were at the position 43 of Nsp16, this site was very close to the conserved catalytic site position 46. Though the revertants R4, R5, R6 and R7 had the mutation at position 43 (Asn to Ser) plus another mutation within Nsp16. There was no reason to attribute these additional changes to the revertant phenotype, especially as they were all well away from catalytic positions. The revertants R3 had a single mutation at Nsp16 postion 130, this was a catalytic residue which formed the conserved catalytic tetrad (K-D-K-E)[21, 23]. Research on West Nile virus (WNV) showed all residues within the K-D-K-E tetrad of the WNV MTase were essential for 2'-O methylation activityl residue D was more critical than other tetrad residues, mutants with a mutation at position 146 (D to E) were viable but exhibited small plaques[27].
Together, these sequencing results identified the single mutation at Nsp16 position 43 (Asn to Ser) or position 76 (Lys to Glu) and position 130 (Asp to Asn) had the ability to revert from the ts mutant Wu"-ts18(cd) to non-ts phenotype, and formed large wild-type plaques; an additional independent mutation in Nsp13 (superfamily 1 helicase) at position 115 (Thr to Ile) could also play an important role on plaque size, being associated with the small plaque phenotype. To date, this is the first report of a Nsp12 ts mutant virus and this mutant can be used to study the role of Nsp13 in MHV virus replication.
Plaque assay of the mutant virus Wu"-ts18(cd) at different temperature
Replication analysis of ts mutant Wu" -ts18 (cd) in 17Cl-1 cells
Sequence analysis of the revertant mutants of Wu"-ts18 (cd)
-
TS mutants and revertants had been described for a large number of positive-strand RNA viruses [5, 11, 14, 17, 33], the major advantage of using ts mutants is that they could be analyzed by temperature-shift and reversion mutation. Generally, the lose or gain of some viral functions after the temperature shift indicates that change of the amino acid in the mutant had affected the formation or activity of the replication-transcription complex. In this study, we tried to identify the locus which was important in the function of Nsp16. Firstly, we constructed the ts mutant which was predicted to have the same phenotype as the Wu"-ts18. The Wu"-ts18 is a reported ts mutant of wild-type MHV-A59 which fails to form plaques or synthesize viral RNA when infection is initiated and maintained at the non-permissive temperature, and produces smaller plaques than the wt virus at 37℃; the ts defects responsible for Wu"-ts18 RNA-negative phenotype are caused by a single point mutation at Nsp16 position 12. In order to limit the reversion to wild-type and further stabilize the mutation locus, three mutations were introduced by altering the amino acid codon of Nsp16 position 12. Our characterization experiments showed that it clearly mimicked the ts phenotype reported for Wu"- ts18. Sequencing results of constructed Wu"-ts18 (cd) confirmed that this single amino acid replacement was sufficient to generate this ts phenotype.
Although the exact function of Nsp16 has not yet been determined, it has been predicted to be a S-adenosyl-Lmethionine (AdoMet)-dependent RNA 2'O-MTase[9, 12, 16] and RNA cap 2'O-MTase activities have previously been identified for various other positive and negative ssRNA viruses, such as Dengue virus[3] and VSV [6, 12, 13]. For WNV MTase, it has been shown that mutations abolishing the 2'O-Mtase activity had a detrimental effect on the replication of a luciferase expressing RNA replicon[27]. Mutagenesis analysis of the feline CoV (FCoV) Nsp16 showed the MTase activity was abolished or severely reduced by all mutations affecting the putative K-D-K-E catalytic tetrad indicating that the structure-based domain of Nsp16 was functionally essential.
Among the ts revertants we obtained in this study, identification and confirmation of three independent second-site mutations of revertants (N43 to S, K76 to E, D130 to N) indicated that the restoration of Nsp16 function could occur in more than one manner. Interestingly, seven of nine revertants had the same mutation at Nsp16 position 43 (N to S), and position 43 was very close to the conserved catalytic site 46 (K), our results indicate that Nsp16 proteins possess economical mechanisms to respond to adverse conditions (such as high temperature). The same type of subtle modifications at position 43 allowed rapid reversion from a nonfunctional ts mutants to functional virus. The other two revertants had a single amino acid change at Nsp16 position 76 (K to E) or postion 130 (D to N). All three single amino acid mutants (R1, R2 and R3) changed the charge of the protein, teplacing a Lys with a Glu at position 76, as in revertant R2, adding a negative charge to the protein, and the proximity was important in stablizing the structure of the protein[21-23].
The sequencing and reverse genetic studies presented in this paper argued that the complete genome sequencing and recapitulation of ts mutations using reverse genetic strategies would be critical for addressing ts mutant viruses in the future. The results of this study provide new insights into the identification of novel residues that could be very important for the functional replication-transcription complex of the coronaviruses.













 DownLoad:
DownLoad: