HTML
-
Studies of the coevolutionary dynamics between Pseudomonas fluorescens SBW25 and bacteriophage Ф2 can explore host resistance and parasite infectivity with applications in the ecological and therapeutic fields. Coevolutionary dynamics determine the efficacy of phage-based therapy. In the study described here, bacterial resistance and phage infectivity fluctuated with culture time, perhaps resulting from random mutation and temporal adaptation, which reminds us of the necessity to consider evolutionary mechanisms when applying phage to treat bacterial infections.
In order to combat the emergence of multidrug-resistant bacteria, phage-based therapy has gained renewed interest (Bradbury J, 2004), although it has not progressed significantly after nearly 100 years of research, possibly because of some inconsistent results that phage did not succeed in controlling bacterial numbers (Horne M T, 1970; Levin B R, et al., 1996). A therapeutic phage with the ability of self-reproduction does not represent a conventional stable medicine, but rather an organism that can interact and evolve with its host. Therefore, the coevolution between phage and bacterium is a powerful determinant of phage-based therapy.
Pseudomonas fluorescens SBW25 and its phage Ф2 have become a model for the study of coevolution between bacteria and phage (Buckling A, et al., 2006; Gómez P, et al., 2011). Our experiments were designed according to previous reports, with some modifications (Buckling A, et al., 2002; Lopez-Pascua L D C, et al., 2008). P. fluorescens SBW25 and its bacteriophage SBW25 Ф2, which were provided by Dr. Lin Jiang (School of Biology, Georgia Tech, Atlanta, GA 30332, USA), were used for these experiments. Cultures were propagated in static and shaking microcosms in 25 ml glass tubes containing 6 mL of standard King's medium B (KB); shaking (where applied) was at 200 rpm, incubation was at 28 ℃. At the beginning of culture, five replicate microcosms were inoculated with 108 cells of P. fluorescens isolate SBW25 and 105 particles of phage SBW25 Ф2. Sixty microliters of each culture were transferred to a fresh microcosm (containing 6 mL KB medium) every 2 days for a total of 12 transfers (24 days).
We used a serial dilution method to count the total number of colony forming units (CFUs) to determine the number of bacteria in a given population, using KB agar plates. Phage particles mixed with a very large number of ancestral P. fluorescens SBW25 cells were spread on a KB agar plate to determine the number of bacteriophage, counted by plaque forming units (PFUs) (Moe J B, et al., 1981). Bacterial resistance or sensitivity, as well as phage infectivity or non-infectivity, were used to evaluate the coevolutionary dynamics between P. fluorescens SBW25 and phage Ф2, as in the reports of Buckling A et al. (2002), in which 20 independent bacterial colonies were streaked across a perpendicular line of phage previously streaked on a KB agar plate. A bacterial colony was defined as resistant if there was no inhibition of growth, otherwise it was defined as sensitive; phage was defined as non-infective when it could not inhibit the bacterial growth, otherwise it was defined as infective.
We measured the resistances of bacterial populations to 'ancestral' phage (day 0), 'middle-transferred' phages (day 2 and day 12), and 'final-transferred' phage (day 24), in order to evaluate the variation of phage infectivity. In the same way, the infectivities of phage populations to ancestral bacteria (day 0), 'middle-transferred' bacteria (day 2 and day 12), and 'final-transferred' bacteria (day 24) were tested, in order to evaluate the variation of bacterial resistance.
Bacterial density was expressed as log10(Nb)/mL [Nb = mean ± SE; Nb (CFU/mL) represents the average number of bacteria per mL from five replicates]. Phage density was expressed as log10(Np+1) [Np = mean ± SE; Np (PFU/mL) represents the average number of phage per ml from five replicates]. Phage infectivity (Nv) and bacterial resistance (Nr) were expressed as the proportional value of inhibited bacterial strips/20 and uninhibited bacterial strips/20, respectively, from five replicates [Nv = (mean ± SE)/20, n = 5; Nr = (mean ± SE)/20, n = 5]. SPSS 17.0 for Windows with one-way ANOVA was used for statistical analyses. Analysis of variance was used to find significant difference at p = 0.05.
The densities of bacteria cultured with phage were lower than those of the phage-absent groups at most time points, but occasionally they were higher (Figure 1A). In static culture at day 22, the bacterial density of the phage-containing group was significantly higher than that of the phage-absent group (p = 0.03); in shaking cultures at days 4 and 24, the phage-containing group had significantly higher bacterial density than the phage-absent group (p = 0.006 and p = 0.043 respectively). After phages have eliminated most of the sensitive cells, genetic resistance can rise, allowing an increase in bacterial densities so as to reach high levels after a period of time (Bull et al., 2014). Static or shaking conditions did not significantly influence the bacterial densities in either the phage-containing group (p = 0.382) or the phage-absent group (p = 0.096).
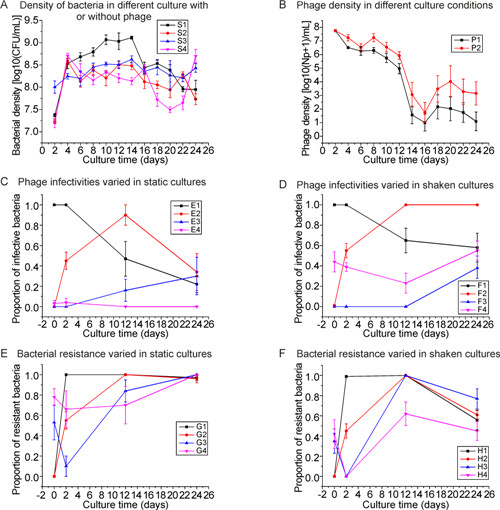
Figure 1. (A) Bacterial density in serial cultures. S1: density of bacteria in static culture without phage; S2: density of bacteria in static culture with phage; S3: density of bacteria in shaken culture without phage; S4: density of bacteria in shaken culture with phage. (B) Phage density in serial cultures. P1: Culture in static conditions; P2: Culturein shaken conditions. (C) Phage infectivity in static culture. E1: 0-day, 2-day, 12-day and 24-day phage inhibition of ancestral (0-day) bacteria; E2: 0-day, 2-day, 12-day and 24-day phage inhibition of 2-day bacteria; E3: 0-day, 2-day, 12-day and 24-day phage inhibition of 12-day bacteria; E4: 0-day, 2-day, 12-day and 24-day phage inhibition of 24-day bacteria. (D) Phage infectivity in shaken culture. F1: 0-day, 2-day, 12-day and 24-day phage inhibition of ancestral (0-day) bacteria; F2: 0-day, 2-day, 12-day and 24-day phage inhibition of 2-day bacteria; F3: 0-day, 2-day, 12-day and 24-day phage inhibition of 12-day bacteria; F4: 0-day, 2-day, 12-day and 24-day phage inhibition of 24-day bacteria. (E) Bacterial resistance in static culture. G1: 0-day, 2-day, 12-day and 24-day bacterial resistance to ancestral (0-day) phage; G2: 0-day, 2-day, 12-day and 24-day bacterial resistance to 2-day phage; G3: 0-day, 2-day, 12-day and 24-day bacterial resistance to 12-day phage; G4: 0-day, 2-day, 12-day and 24-day bacterial resistance to 24-day phage. (F) Bacterial resistance in shaken culture. H1: 0-day, 2-day, 12-day and 24-day bacterial resistance to ancestral (0-day) phage; H2: 0-day, 2-day, 12-day and 24-day bacterial resistance to 2-day phage; H3: 0-day, 2-day, 12-day and 24-day bacterial resistance to 12-day phage; H4: 0-day, 2-day, 12-day and 24-day bacterial resistance to 24-day phage.
Figure 1B shows that phage density under shaking conditions was significantly higher than that under static conditions (p = 0.02). Although bacterial density in static cultures was significantly higher than that in shaken cultures at day 20 (p = 0.018), the phage density in static culture (based on the PFU number) was still significantly lower than that in shaken cultures at this time point (p = 0.027). These results indicated that spatially heterogeneous environments (a static microcosm) can promote rapid genotype diversification (Brockhurst M A, et al., 2006), resulting in phage differentiation in response to their genotypically diversified hosts. Therefore, there were lower numbers of PFU in the static group when ancestral bacteria (with less differentiation) were used as indicators. From day 2 to 16, phage density declined from ~107 to ~10 PFU/mL in the static group. Moreover, under static conditions, phage densities on some subsequent days (day 12, day 14, day 16, day 22 and day 24) were significantly lower than on day 2 (Figure 1B). There were no significant differences in phage densities under shaking conditions compared with static conditions.
Figure 1C shows that in static cultures, the infectivity of 2-day, 12-day and 24-day phage to ancestral bacteria declined significantly with time. Thus, phage infectivity did not increase with prolongation of the incubation time. When 2-day bacteria were used as indicators, phage infectivity increased significantly from 0 to 12 days, and then declined significantly between 12 days and 24 days. However, when 12-day bacteria were used as the tested strain, only 24-day phage had a significantly higher infectivity than 0-day and 2-day phages. When 24-day bacteria were used as the tested strain, there was no significant difference in phage infectivities. In Figure 1D, which shows the results with shaken cultures, phage infectivity to ancestral bacteria declined significantly over time and showed a similar pattern to that with static ancestral bacteria (compare Figures 1C and 1D, curves E1 and F1). When 2-day bacteria were tested, phage infectivity increased significantly from days 0 to 12 and then remained constant at day 24. When 12-day bacteria were used as indicators, phage infectivity increased significantly from day 12 to day 24. Phage infectivity was significantly different between day 0 and day 12, and between day 12 and day 24, when 24-day bacteria were used as indicators.
Figure 1E, which presents data from static cultures, shows that when ancestral phages were used as indicators, bacterial resistance increased significantly from day 0 to day 2, and then did not vary significantly from day 2 to day 24. When 2-day phages were used as indicators, bacterial resistance increased significantly until day 12 and then remained constant; when 12-day phages were used as indicators, bacterial resistance declined significantly from day 0 to day 2, and then increased significantly from day 2 to day 12; finally, when 24-day phages were used, only 24-day bacteria showed a significantly higher resistance than 2-day and 12-day bacteria. Figure 1F shows data for shaken cultures. When ancestral phages were used as indicators, bacterial resistance varied significantly, except between day 2 and day 12; when 2-day phages were used as indicators, bacterial resistance increased significantly from day 0 to day 12, and then declined significantly from day 12 to day 24; when 12-day phages were used, bacterial resistance varied significantly; and lastly, with 24-day phages, 2-day bacteria displayed significantly lower resistance than those at days 0, 12 and 24.
Our results showed that phage did not always control bacterial density effectively, which was consistent with the findings of Wei Y et al. (2010), but inconsistent with the report of Betts A et al. (2013). Engineered phages, phage cocktails and phage-antibiotic combinations can control bacterial density for certain lengths of time (Reardon S, 2014; Schmerer M, et al., 2014; Torres-Barceó C, et al., 2014), but these strategies just delay the evolution of phage-resistant bacteria (Lu T K, et al., 2011). Eventually, bacteria with host defense systems would be driven to evolve to escape phage attack (Clokie M R, et al. 2011; Bull J J, et al., 2014).
Our findings did not conform to the pattern that bacterial resistance and phage infectivity can be increased in a constant arms race of coevolution (Buckling A, et al., 2002). Bacterial resistance and phage infectivity varied with the conditions. These variations in host resistance and phage infectivity might reflect the fact that random mutation and temporal selection each influence the characteristics of this coevolutionary process. Many mutations are random, not directed (Lederberg J, et al., 1952). Both the phage and its host experience the same mechanisms of evolution–random mutation and temporal selection.
Reports of evolution being driven by directional selec-tion to develop more resistant bacteria and more infective phage, and of the evolutionary 'training' of therapeutic phages to combat bacterial infections, lead to a significant question (Buckling A, et al., 2002; Betts A, et al., 2013). Can this resistant/infective capacity increase forever? From the viewpoint of evolution, such an outcome is incredible. In nature, random mutation is the driver of evolution. Directional selection for more virulent phage may fail. In our experiments, phage did not control bacterial density significantly (Figure 1A).
In a large population, there are many genotypes (resulting from random mutation) ahead of host-parasite interactions, that is to say, there are many genotypes that are pre-existing, quite apart from the influence of host-parasite interactions. Bacteria-phage adaptation is just a transient state due to the unceasing random mutations that occur in both bacterial and phage populations. Therefore, host resistance and parasite infectivity is a temporal, genotype-specific interaction. The temporal selective dynamics of host-parasite coevolution produces variations in host resistance and parasite infectivity. Parasites have no evolutionary advantage over their hosts, and their infectivity always varies (Forde S E, et al., 2008). Bacteria can develop resistance to phages through a variety of different mechanisms, which can have both genetic and non-genetic causes (Lu T K, et al., 2011; Bull J J, et al., 2014).
Therefore, much more work should be carried out to uncover the mechanisms of resistance and infective variation and to improve our understanding of phage-based therapy. Although the effectiveness of some phage-based therapies seems excellent, systematic proof is required to demonstrate that phage-based therapies that appear promising are in fact reliable and reproducible.
-
This work was supported by the Open Funding Project of the Engineering Research Center of Eco-environment in Three Gorges Reservoir Region-Ministry of Education (KF2013-07). We gratefully thank Dr. Lin Jiang, Dr. Yin Chen, Jiaqi Tan and Zhichao Pu for their help. All the authors declare that they have no competing interests. This article does not contain any studies with human or animal subjects performed by any of the authors.













 DownLoad:
DownLoad: