HTML
-
Ebola virus and Marburg virus are filamentous, enveloped, single-stranded, negative-sense RNA viruses belonging to the family Filoviridae. Serological and genetic analysis have enabled the distinction of five species of the genus Ebolavirus: Ebola virus, Sudan virus, Taï Forest virus, Bundibugyo virus, Reston virus, and two species of the genus Marburgvirus: Marburg virus and Ravn virus (Kuhn J, et al., 2013). Furthermore, there is a newly discovered filovirus named Lloviu virus assigned to the genus Cuevavirus, with one species, Lloviu cuevavirus (Negredo A, et al., 2011; Kuhn J, et al., 2013).
The genomic structures of all these viruses are very similar; they are approximately 19 kb in length, containing seven genes arranged sequentially in the order: nucleoprotein, viral protein (VP35), matrix protein (VP40), glycoprotein, replication-transcription protein (VP30), minor matrix protein (VP24), NA-dependent RNA polymerase (L). EBOV also expresses secreted nonstructural glycoprotein sGP, ssGP and delta-peptide (Sanchez A, et al., 2007).
Ebola virus and Marburg virus are causative agents of severe hemorrhagic fever in humans and non-human primates, with high mortality rates, affecting 50%–90% of infected individuals (Feldmann H, et al., 2003). Transmission of filovirus is generally by direct contact with blood, secretions or infected tissues, although experiments on monkeys provided evidence of an airborne route of infection (Jaax N, et al., 1995).
Since the discovery of Marburg virus disease in Germany in 1967, sporadic outbreaks of Marburg and Ebola virus disease have been reported from different countries in central Africa (Feldmann H, et al., 2003). Since December 2013, there has been a new outbreak of Ebola virus disease caused by Ebola virus infection; this started in a remote rural part of Guinea and then spread through Guinea, Liberia, Sierra Leone, and Nigeria in western Africa. As of 26 September 2014, 6553 (probable, confirmed and suspected) cases and 3083 deaths have been reported and the increase in cases continues to accelerate in these countries, with widespread and intense transmission (WHO, 2014). On 8 August 2014, the World Health Organization announced the current outbreak in western Africa as an international public health emergency. The global epidemic tendency remains ambiguous to date and the increasing possibility of the virus invading other continents is receiving more attention worldwide. For the prevention of Ebola virus transmission, the reliable early detection of Ebola virus infection is essential. Although some RT-PCR detection techniques have been developed to screen for Ebola virus and Marburg virus (Huang Y, et al., 2012), no filovirus-specific antibody-based indirect enzyme-linked immunosorbent assays (indirect ELISAs) for serological diagnosis are available in China.
Filovirus nucleoprotein may be the ideal target antigen because of its abundance in filovirus particles and its strong antigenicity (Niikura M, et al., 2001, 2003; Bharat T, et al., 2012). Nucleoprotein consists of 739 or 695 amino acid residues, with a conserved hydrophobic N-terminus and a variable hydrophilic C-terminal part (Niikura M, et al., 2001; Sanchez A, et al., 2007; Lötfering B, et al., 1999). It plays an important role in the replication of the viral genome and is essential for formation of the nucleocapsid (Watanabe S, et al., 2006). It has been demonstrated that the C-terminal half of nucleoprotein has strong antigenicity (Saijo M, et al., 2001). The conformational and linear epitopes of nucleoprotein have been identified for several Ebola viruses (Ikegami T, et al., 2003; Niikura M, et al., 2001, 2003).
In this study, we developed a specific ELISA using recombinant Ebola virus nucleoproteins and Marburg virus nucleoproteins to detect immunoglobulin G (IgG) antibodies to Ebola virus and Marburg virus. This ELISA system will be useful for virus disease epidemiological and virological surveillance.
-
Plasma samples were collected from individuals passing through the Guangdong port of entry and were offered by Guangdong Inspection and Quarantine Technology Center.
-
The DNA encoding the full-length nucleoprotein was amplified by PCR from the synthetic nucleoprotein genes of Ebola virus H.sapiens-tc/COD/1976/Yambuku-Mayinga (Ebola virus, RefSeq #NC_002549) and Marburg virus H.sapiens-tc/KEN/1980/Mt. Elgon-Musoke (Marburg virus, RefSeq #NC_001608) using the following primers: EBOV-ZNP-1F (5′-GGA TCC ATG GAT TCT CGT CCT CAG AAA AT-3′), EBOV-ZNP-1R (5′-GTC GAC TCA CTG ATG ATG TTG CAG GAT T-3′) and MARV-NP-2F (5′-CAT ATG ATG GAT TTA CAC AGT TTG TTG GAG T-3′), MARV-NP-1R (5′-GTC GAC CTA CAA GTT CAT CGC AAC ATG TCT-3′) and cloned into pGEM®-T easy vector (Promega, Madison, USA). After sequence confirmation, the Ebola virus nucleoprotein gene was digested with Bam HI and Sal Ⅰ, purified and subcloned into the prokaryotic expression pET-28a vector (Novagen, Cambridge, USA). The Marburg virus nucleoprotein gene was digested with Nde Ⅰ and Sal Ⅰ, purified and subcloned into a pET-28a vector. These His-tagged nucleoproteins were expressed in E. coli (BL21 strain). Both pCMV-ENP-His and pCMV-MNP-His were constructed using standard genetic engineering techniques, as above. The Ebola virus nucleoprotein was excised from pGEM®-T easy vector (Promega) using Bam HI and Sal Ⅰ and subsequently cloned into the pCMV-His (Beyotime, Shanghai, China). The resulting plasmid was termed pCMV-ENP-His. Similarly, the Marburg virus nucleoprotein was excised from pGEM®-T easy vector (Promega) using Eco RI and Sal Ⅰ and subsequently cloned into the pCMV-His (Beyotime). The resulting plasmid was termed pCMVMNP-His.
-
Small-scale pilot experiments were performed for each recombinant pET-28a construct in order to determine optimal bacterial expression conditions for His-tagged Ebola virus nucleoprotein and Marburg virus nucleoprotein. Briefly, 5 mL shaker flask cultures of transformed E. coli were grown in lysogeny broth at 37 ℃ to an A600 = 0.5–0.6. Each culture was split into three flasks and induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) to final concentrations of 0.1, 0.4 and 1.0 mmol/L at different temperatures. Cultures were then grown under induction conditions for 3 h. Subsequently, soluble fractions and insoluble pellets were prepared by centrifugation from whole cell lysates of E. coli transformed with pET-28a-based vectors and then analyzed by SDS-PAGE. Using optimal temperature and IPTG concentrations, as determined by these studies, a time-course investigation was carried out to further maximize total fusion protein yields. Analysis by SDS-PAGE was performed on the soluble fraction and insoluble pellet from the harvested samples after induction.
-
To generate and purify His-tagged nucleoprotein, a 100 mL shaker flask culture of pET-28a-EBOV-NP or pET-28a-MARV-NP transformed BL21 (DE3) cells was grown in lysogeny broth containing 50 mg/mL kanamycin to an A600 = 0.5–0.6 at 37 ℃ and then induced with a final IPTG concentration of 0.4 mmol/L. After incubation at 37 ℃ or 16 ℃ for 3 h or 8 h, cells were harvested by centrifugation at 10, 000 × g for 10 min at 4 ℃ (BECKMAN COULTER Avanti J-26XP). The cell pellet was frozen at -20 ℃ and subsequently thawed and resuspended in BugBuster reagent (Novagen) by pipetting or gentle vortexing. The cell suspension was incubated on a shaking platform or rotating mixer at a slow setting for 10–20 min at room temperature. Insoluble cell debris was removed by centrifugation at 16, 000 × g for 20 min at 4 ℃ (Thermo Scientific Sorvall Legend Micro17R). The remaining soluble extract was loaded directly onto Novagen His·Binding purification resin (Ni-charged) according to the manufacturer's instructions (Novagen). Purified nucleoprotein was aliquoted and stored at -20 ℃.
-
Rabbit polyclonal antibody was prepared by subcutaneous injection of purified nucleoprotein mixed with an equal volume of complete Freund's adjuvant and boosted subcutaneously three times with recombinant nucleoprotein mixed with an equal volume of incomplete Freund's adjuvant in a total volume of 1 mL protein-adjuvant mixture for each injection. The specific antibody was purified using PureProteomeTM Protein A Magnetic Beads according to the manufacturer's instructions (Millipore). The titer of purified antiserum was >1:4000 in a Western blotting assay, in which nucleoprotein-expressing cells were used as the antigen.
-
293T cells were transfected with pCMV-C-His, pCMV-ENP-His or pCMV-MNP-His plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses were performed with a 10% acrylamide gel and an Immobilon PVDF membrane (Millipore) respectively. The rabbit anti-nucleoprotein polyclon al antibody, as primary antibody, was used at a dilution of 1:4000. The bound antibody was detected with alkaline phosphatase -conjugated goat anti-rabbit IgG (H+L) polyclo nal and BCIPNBT substrate (Beyotime). Reactions were stopped by immersing developed membranes in water, followed by immediate high resolution scanning for permanent recording. A monoclonal antibody to His-tag was purchased from Abmart (Shanghai).
-
ELISA plates (Costar) were coated with the nucleoprotein antigens (50 ng of Ebola virus nucleoprotein and 10 ng of Marburg virus nucleoprotein dissolved in 100 μL sodium carbonate-sodium bicarbonate buffer per well) or control antigens in phosphate buffered saline (PBS) at 4 ℃ overnight and then washed with PBS containing 0.05% Tween 20 (PBST). Unspecific binding of the antibodies was avoided by blocking with 5% skim milk (100 μL/well) for 2 h at room temperature. After washing three times with PBST, 100 μL of appropriately diluted serum or plasma sample in PBST containing 2% skim milk was added and incubated for 1 h at room temperature. After washing three times with PBST, the bound antibodies were detected by using the following secondary antibodies conjugated with horseradish peroxidase diluted in 2% skim milk in PBST: goat anti-rabbit IgG (Beyotime)or goat anti-human IgG (Boster). After incubation for 1 h at room temperature and three PBST washes, 200 μL of 3, 3′, 5, 5′-tetramethylbenzidine (Sigma) was added to each well and the mixture was incubated for 15 min at room temperature. The reaction was stopped by adding 1 N sulfuric acid to the mixture, and the opt ical density at 450 nm was measured (Bio-RAD iMarkTM microplate reader).
Plasma samples
Plasmid construction
Optimization of recombinant nucleoprotein expression in bacteria
Purification of recombinant soluble nucleoprotein
Development of rabbit anti-nucleoprotein polyclonal antibody
Recombinant nucleoprotein expression in mammalian cells
Western blotting
ELISA
-
The complete nucleoprotein genes of Ebola virus and Marburg virus were subcloned into the pET-28a plasmid in-frame with the six histidine codons which represent the N-terminal sequence of the expressed fusion protein (Figure 1). The expected sizes of the Ebola virus nucleoprotein and Marburg vir us nucleoprotein are 104 kDa and 96 kDa, respectively. After induction with IPTG, bacteria harboring pET-28a-EBOV-NP or pET-28a-MARV-NP, a polypeptide of about 100 kDa was synthesized; this polypeptide was not detected in non-induced or control cultures (Figure 2A). For Ebola virus nucleoprotein, vector pET-28a and E. coli BL21 (DE3) cells were suitable for higher level of soluble protein expression, with induction performed using 0.4 mmol/L IPTG at 37 ℃ for 3 hours (Figure 2A). These conditions produced ~8 mg Ebola virus nucleoprotein fusion protein per liter of shake flask culture (Figure 2B). However, the same conditions did not result in a similar yield of soluble Marburg virus nucleoprotein but amount of occlusion bodies (Figure 2C).
Figure 1. Recombinant expression constructs identification. A. Restriction endonuclease digestion for pET-28a-EBOV-NP confirmation; B. Restriction endonuclease digestion for pET-28a-MARV-NP confirmation.
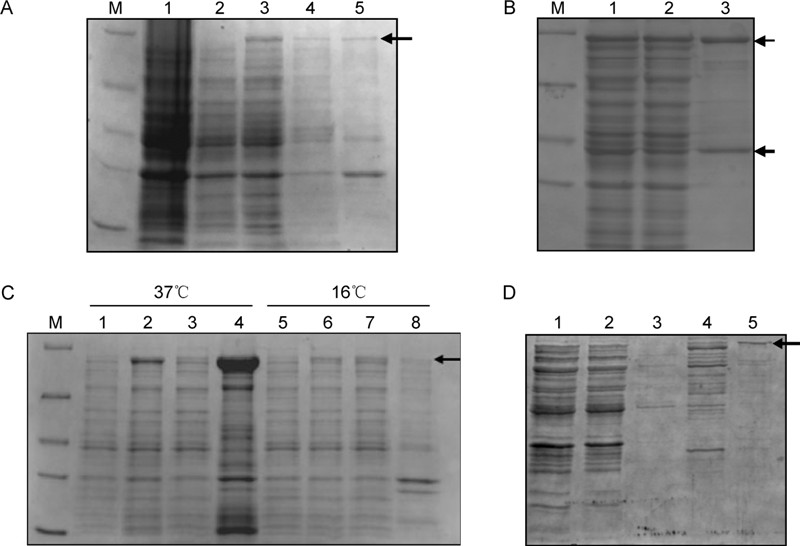
Figure 2. Expression of recombinant Ebola virus nucleoproteins and Marburg virus nucleoproteins in bacteria. A. SDS-PAGE of proteins in whole-host bacterial cell lysate (lane 1), non-induced host cell containing pET-28a-EBOV-NP (lane 2), induced host cell containing pET-28a-EBOV-NP (lane 3), supernatant of induced host cell containing pET-28a-EBOV-NP after BugBuster reagent treatment (lane 4), insoluble fraction of induced host cell containing pET-28a-EBOV-NP after BugBuster reagent treatment (lane 5). B. SDS-PAGE of proteins in supernatant of whole bacterial cell lysate (lanes 1 and 2), His Binding resin capture Ebola virus nucleoprotein eluate (lane 3). C. SDS-PAGE of proteins in whole-host bacterial cell lysate (lanes 1 and 5), induced host cell containing pET-28a-MARV-NP (lanes 2 and 6), supernatant of induced host cell containing pET-28a-MARV-NP (lanes 3 and 7) after BugBuster reagent treatment, insoluble fraction of induced host cell containing pET-28a-MARV-NP (lanes 4 and 8) after BugBuster reagent treatment at 37 ℃ and 16 ℃. D. SDS-PAGE of proteins in supernatant of whole bacterial cell lysate (lanes 1 and 2), wash buffer (lanes 3 and 4), and His Binding resin capture of Marburg virus nucleoprotein eluate (lane 5).
To obtain a sufficient concentration of soluble Marburg virus nucleoprotein fusion protein for our studies, pilot experiments were performed to determine parameters for optimal fermentation, including criteria for appropriate growth temperature, IPTG concentration and time of harvest following induction. For optimal expression of Marburg virus nucleoprotein fusion protein, pET-28a-MARV-NP transformed BL21 (DE3) cells were induced with 0.4 mmol/L IPTG at 16 ℃ for 8 h (Figure 2C). These conditions resulted in a 1.5-fold increase compared with initial studies of soluble protein yield, ≈6.5 mg of MBPnucleoprotein fusion protein per liter of shake flask culture grown in lysogeny broth (Figure 2D).
The expression of Ebola virus nucleoprotein was high, whereas the expression of Marburg virus nucleoprotein was relatively low. The expressions of soluble Ebola virus nucleoprotein and Marburg virus nucleoprotein in bacteria were confirmed by immunoblotting using anti-His monoclonal antibodies. These recombinant nucleoproteins were purified, as described in the Materials and Methods section. The immunological reaction between all recombinant nucleoproteins expressed in bacteria and mammalian cells and between rabbit anti-nucleoprotein polyclonal antibody were detected by SDS-PAGE and immunoblotting, respectively (Figure 3). Although the purified Marburg virus nucleoprotein was weak, it was demonstrated by Western blotting (Figure 3D). These purified nucleoproteins were used as antigens for the ELISA described in the following experiments.
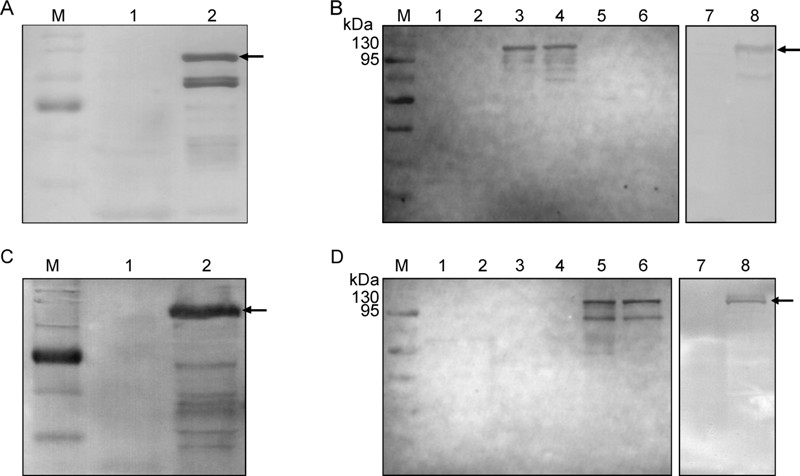
Figure 3. Demonstration of expressed Ebola virus nucleoprotein and Marburg virus nucleoprotein by Western blotting. A. Bacteria harboring pET-28a (lane 1) or pET-28a-EBOV-NP (lane 2) synthesized protein detection using the monoclonal antibody to His-tag. B Protein analysis of lysates from uninduced or induced bacteria harboring pET-28a (lanes 1 and 2), pET-28a-EBOV-NP (lane 3), pET-28a-MARV-NP (lane 5), purified Ebola virus nucleoprotein (lane 4) or Marburg virus nucleoprotein (lane 6) and 293T cells transfected with pCMV-C-His (lane 7) or pCMVENP-His (lane 8) using purified rabbit polyclonal antibody against Ebola virus nucleoprotein. C. Bacteria harboring pET-28a (lane 1) or pET-28a-MARV-NP (lane 2) synthesized protein detection using the monoclonal antibody to His-tag. D. Protein analysis of lysates from uninduced or induced bacteria harboring pET-28a (lanes 1 and 2), pET-28a-EBOV-NP (lane 3), pET-28a-MARV-NP (lane 5), purified Ebola virus nucleoprotein (lane 4) or Marburg virus nucleoprotein (lane 6) and 293T cells transfected with pCMV-C-His (lane 7) or pCMV-MNP-His (lane 8) using purified rabbit polyclonal antibody against Marburg virus nucleoprotein.
Immunoblotting confirmed the identity of the polypeptide and revealed the synthesis of a related protein that migrated slightly more quickly (Figure 3). This related protein seems to be a degradation product and levels could be significantly reduced when higher concentrations of protease inhibitors were present in the lysis buffer (Prehaud C, et al., 1998).
-
Preliminary tests were conducted by coating wells with increasing amounts of recombinant proteins and using anti-nucleoprotein rabbit serum. A strong signal (A450 > 1) was observed with 100 ng (or more) of the purified Ebola virus nucleoprotein and 20 ng (or more) of the Marburg virus nucleoprotein but this was reduced to the background level with 50 ng Ebola virus nucleoprotein and 10 ng Marburg virus nucleoprotein. It was then decided to coat the wells with 50 ng or 10 ng recombinant purified proteins. A high background signal was a common problem in indirect ELISA, so the plate should be blocked. In our study, 5% skim milk seemed to be better at reducing the background signal to a low level than 2% bovine serum albumin. Under these optimal conditions, a better reactivity to Ebola virus nucleoprotein and Marburg virus nucleoprotein were shown when serial five-fold dilutions of the antibodies (1:100 to 1:312, 500, 10-4 to 101 μg/mL)were prepared (Figure 4A and 4B). The detection limit for specific antibodies using this assay was approximately 0.01 to 0.1 μg/mL.
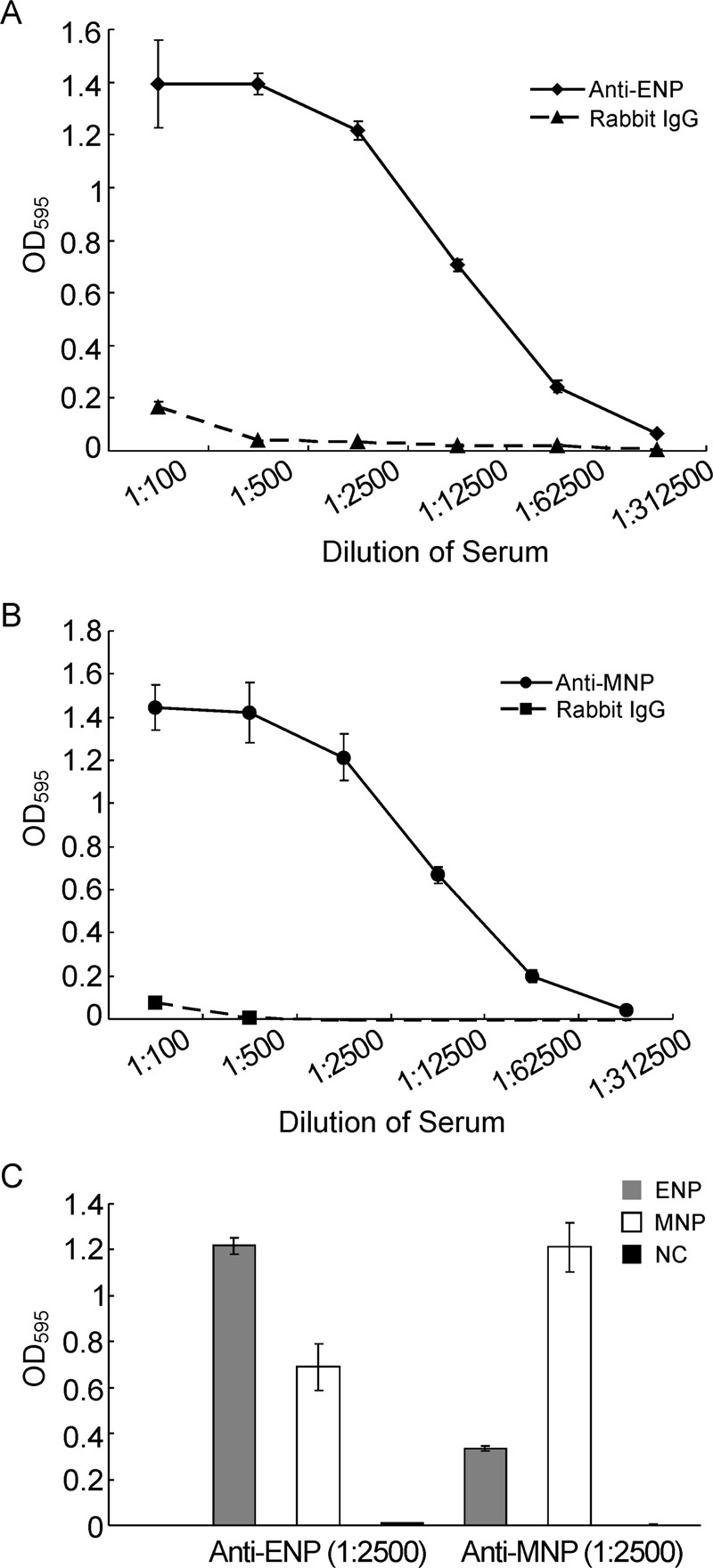
Figure 4. Sensitivity and specificity of ELISAs. His-nucleoproteins were used as antigens. Serial five-fold dilutions of rabbit serum to Ebola virus nucleoprotein, Marburg virus nucleoprotein, and rabbit IgGs (used as negative control antibody) were incubated in Ebola-nucleoprotein (A) or Marburg virus nucleoprotein coated plate (B). C. Diluted rabbit serum (1:2500) was incubated in Ebola virus nucleoprotein and Marburg virus nucleoprotein coated plates simultaneously; significant differences in optical density are shown between the homologous antigen and other antigens. Standard deviations for three repeats of each dilution are shown. ENP, Ebola virus nucleoprotein; MNP, NC, Negative control.
-
The specificity of the ELISA was assessed by testing the antisera of rabbit immunized with Ebola virus nucleoprotein and Marburg virus nucleoprotein. We found that Ebola virus nucleoprotein specific IgG was clearly detected in these rabbit antisera (Figure 4C). All the anti-Ebola virus IgG antibodies in the sera showed low reactivity to Marburg virus nucleoprotein. Similarly, anti-Marburg virus nucleoprotein serum reacted to Marburg virus nucleoprotein but not to Ebola virus nucleoprotein. These results indicated that this purified nucleoproteins based ELISA sufficiently detected filovirus-specific antibodies.
-
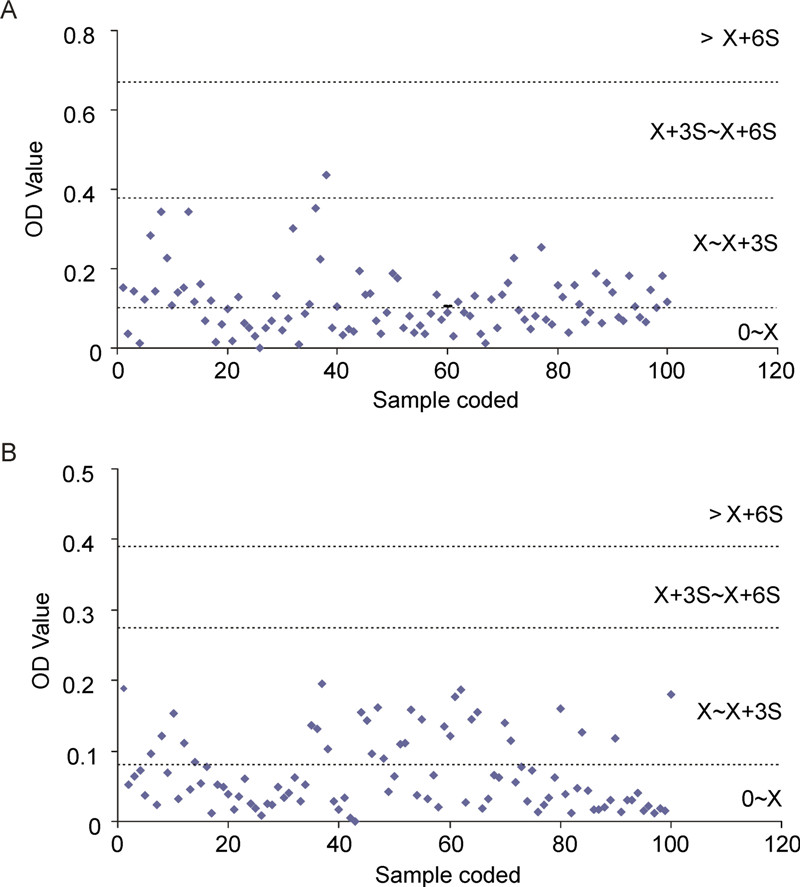
To further confirm the specificity of our ELISA, we used plasma samples collected from individuals passing through the Guangdong port of entry (Figure 5). The cutoff optical density values (i.e., the mean plus three standard deviations of the negative serum samples) were 0.35 and 0.2 for Ebola virus nucleoproteins and Marburg virus nucleoproteins, respectively. According to these thresholds, most of the plasma samples were negative, except for one suspected case, which was positive for Ebola virus nucleoprotein antibody; this was most probably caused by interference by irrelevant proteins in the sample or error in operation and needs to be confirmed by other more sensitive and accurate methods.
Expression and purification of recombinant Ebola and Marburg virus nucleoproteins
Use of recombinant nucleoprotein as antigens for ELISA
Specificity of the nucleoprotein-based ELISA
Analysis of plasma samples in the nucleoprotein-based ELISA
-
Both ELISA and indirect immunofluorescent methods using cells infected with Ebola virus and Marburg virus have previously been developed and used to detect filovirus-specific antibody (Ksiazek T, et al., 1999a; van der Groen G, et al., 1983). These methods require preparation of antigens by handling live Ebola virus and Marburg virus. To make it possible to detect antibodies to Ebola virus and Marburg virus without handling live viruses, increasing number of researchers have developed an IgG ELISA using recombinant Ebola virus and Marburg virus proteins (Saijo M, et al., 2006; Nakayama E, et al., 2010; Ou W, et al., 2011).
In this study, we established a nucleoprotein-based ELISA to detect Ebola virus and Marburg virus specific antibodies. We first expressed nucleoproteins from E. coli and confirmed the antigenicities of the recombinant Ebola virus nucleoproteins and Marburg virus nucleoproteins expressed in bacteria and mammalian cells by Western blotting. The sensitivities and specificities of ELISAs prepared with purified Ebola virus nucleoprotein and Marburg virus nucleoprotein as the antigen were evaluated. Under optimal conditions, a better reactivity to Ebola virus nucleoprotein and Marburg virus nucleoprotein was shown with serial five-fold dilutions of the antibodies (1:100 to 1:312, 500, 10-4 to 101 μg/mL) and the detection limit for specific antibodies using this assay was approximately 0.01 to 0.1 μg/mL, as shown in Figure 4; this limit is similar to results obtained for glycoprotein-based ELISA (Nakayama E, et al., 2010). The same study suggests that if proper antigens and sensitive assays are available, immunoglobulin M (IgM) ELISA can be used to diagnose acute Ebola virus and Marburg virus infections and support antigen capture and reverse transcription-PCR detection, the most commonly used technologies (Nakayama E, et al., 2010).
In addition, we found that IgGs specific for Ebola virus nucleoprotein and Marburg virus nucleoprotein were clearly detected in these rabbit antisera (Figure 4C), although the anti-Ebola virus IgG antibodies in the sera showed low reactivity to Marburg virus nucleoprotein. The explanation for this observation might be that common antibody epitopes seem to be present (Niikura M, et al., 2003), because the nucleoproteins of Ebola virus and Marburg virus contain similar amino acid sequences (Sanchez A, et al., 1992). We further evaluated its ability to detect antibodies to Ebola virus and Marburg virus using human sera samples collected from individuals passing through the Guangdong port of entry offered by Guangdong Inspection and Quarantine Technology Center. We eventually set the threshold at the mean plus three standard deviations of average optical densities of sera tested at a 1:1000 dilution. With this threshold, the ELISA system using these two recombinant nucleoproteins has good sensitivity and specificity.
Despite the discovery of Reston virus in domestic pigs in the Philippines (Barrette R, et al., 2009)and the discovery of fruit bat species as a potential reservoir for Ravn virus and Marburg virus (Leroy E, et al., 2005; Towner J, et al., 2009), the search for reservoirs and potential amplifying hosts remains ongoing. Advanced diagnostic technologies are welcome, and our nucleoprotein-based specific antibody detection ELISA may be a useful tool for future ecological and seroepidemiological studies in areas where the disease is endemic. For the more recent Ebola virus disease outbreak in western Africa, the World Health Organization advises countries to strengthen epidemiological and virological surveillance of Ebola virus disease, reporting human infections as applicable, as well as other national health preparedness and response actions.
In conclusion, we have developed ELISA systems to detect IgG antibodies against Ebola virus and Marburg virus using recombinant nucleoproteins. These ELISA systems can be used to detect antibodies against Ebola virus and Marburg virus in a facility without a BSL-4 laboratory. More sensitive assays are necessary for various specimens. Ksiazek et al. (1999b) reported the limitation of Ebola virus IgG detection for the diagnosis of Ebola virus infection. Some patients with Ebola virus infections died before the IgG response could be detected. For an accurate diagnosis of acute Ebola virus and Marburg virus infections, antigen ELISA, reverse transcription-PCR, isolation of infectious agents and other methods of detecting the virus are necessary. IgM antibodies can also be useful for inferring recent onset of the immune response. ELISA using recombinant glycoprotein antigens is an available diagnostic method to distinguish antibodies specific to the respective species of filoviruses, and can provide a basis for serological classification, owing to the larger genetic variability of this protein (Nakayama E, et al., 2010). Thus, other diagnostic systems, such as a filovirus antigen capture ELISA and an IgM capture ELISA using recombinant nucleoproteins or glycoproteins of filoviruses, are under construction in our laboratory.
-
This work was supported by Important National Science & Technology Specific Projects (2012ZX10004403).
We thank Dr.Shi Yongxia and her colleagues in Guangdong Inspection and Quarantine Technology Center, who offered the plasma samples of individuals passing through the Guangdong port of entry.
-
All the authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
-
HY designed the experiments. HY, ZY and YM carried out the experiments. HY, ZY and ZZ analyzed the data. HY, ZY, SD and YZ wrote the paper. All authors read and approved the final manuscript.













 DownLoad:
DownLoad: