-
Dear Editor,
Aquatic birnaviruses are a group within the family Birnaviridae that comprise isolates from fsh and shellfsh from both fresh-and seawater (Woo and Bruno, 1999). Members of the family Birnaviridae are icosahedral viruses approximately 60 nm in diameter composed of fve polypeptides and two segments of double-stranded RNA (Dixon et al., 2008). In this report, an aquatic birnavirus was isolated from rainbow trout fry, Onhorhynchus mykiss, during a fsh health inspection in fsh-farms in the west region of Ukraine near the rivers Siret and Cheremosh (Chernivtsi region). Preliminary examination of infected fsh revealed a range of lesions, particularly in pancreatic tissue. Morphological changes, such as vacuole enlargement and cell rounding, were caused by viruses in appropriate cell lines. Investigation by electron microscopy demonstrated that the isolated virus was ultrastructurally similar to infectious pancreatic necrosis virus (IPNV). In addition, the nucleotide sequences of the 1120 bp VP2 gene fragments were analyzed and the identity of the isolated IPNV to strain Sp was revealed. The identity of nucleotide sequences was 97%–99%; the isolates of Sp strains most closely related to the Ukrainian IPNV isolate were found in Norway.
During June 2013, a total of 140 fish samples were continuously collected from moribund and dead rainbow trout fry (body weight 4–7 g) in several fish-farms near the rivers Siret and Cheremosh, in the Chernivtsi region of Western Ukraine. Samples of internal organs (pancreas, kidney and spleen) were removed from individual fsh and placed into 1.5 mL microcentrifuge tubes. Samples were transported on ice to the laboratory and processed immediately.
RTG-2, FHM and EPC cell lines were maintained in MEM medium (PAA) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin and 10% foetal bovine serum (FBS Gold, PAA). The samples of internal organs were homogenized with MEM and filtered through 0.45 µm membrane (Sarstedt). Then the virus suspension was inoculated onto 24-hour cell monolayers growing in 25 cm 2 flasks. After absorption for 60 min at 20 ℃, MEM medium supplemented with 2% of FBS was added to the monolayers. When a complete viral cytopathic effect (CPE) was evident, the tissue culture supernatant was harvested and centrifuged at 2500 × g for 10 min at 4 ℃ to remove cell debris. The 50% tissue culture infective dose (TCID50/mL) of the resulting supernatant was determined (Reed and Muench, 1938).
The virus was purifed from tissue culture supernatant by ultracentrifugation. Briefly, after cell debris was separated by centrifugation at 2500 × g for 10 min at 4 ℃ the pellet was discarded and the supernatant was centrifuged in a Beckman L5-50B centrifuge with an SW-40 rotor for 60 min at 70500 × g at 4 ℃. The virus pellet was suspended in TNE (50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1 mmol/L EDTA, pH 7.5) and centrifuged at 2500 × g for 5 min at 4 ℃. Then the virus suspension was used for electron-microscopy investigation and viral RNA extraction. For electron microscopy the viral suspension was stained with 2% uranyl acetate.
Genomic viral RNA was extracted from collected fsh internal organs, virus-infected cell culture supernatant and purified virus suspension using a GeneJETTM RNA Purification Kit (ThermoScientific) as described in the manufacturer's protocol. The cDNA synthesis was conducted using the RevertAidTM Premium First Strand cDNA Synthesis Kit (ThermoScientifc).
The aquatic birnavirus-specific primers PrA forward 5′-AAA GCC ATA GCC GCC CAT GAA C-3′ and PrA reverse 5′-TCT CAT CAG CTG GCC CAG GTA C-3′ were used for amplifcation of a fragment of viral dsRNA targeting the IPNV VP2 protein (Blake et al., 1995). The PCR mixture consisted of 12.5 μL of DreamTaq Green PCR MasterMix (ThermoScientific), 20 pmol of each primer and 1 μL of cDNA, made up in nuclease-free water to a total volume of 25 μL. The amplifcation was conducted with a pre-dwell cycle of 50 ℃ for 15 min, and then an initial cycle of 94 ℃ for 2 min, followed by 35 cycles of 94 ℃ for 30 s, 58 ℃ for 30 s and 72 ℃ for 60 s. A fnal extension step was conducted at 72 ℃ for 7 min. The PCR products were analysed by 2.0% agarose gel electrophoresis.
The PCR products were purified with the Silica Bead DNA Gel Extraction Kit (ThermoScientific, Waltham, USA) and subjected to nucleotide sequence analysis using a 3130 Genetic Analyzer (Applied Biosystems, Waltham, USA). The sequences were aligned with sequences available in the GenBank database (NCBI) using CLUSTAL W software Molecular Evolutionary Genetics Analysis (MEGA) version 6.0. The nucleotide sequence was registered in GenBank under accession number KJ596654.
In this report we describe the isolation of IPNV from rainbow trout, O. mykiss, fry in fish-farms near the Siret and Cheremosh rivers, Chernivtsi region, Western Ukraine. All moribund and dead trout were found on the surface of fish tanks during screening. External signs of moribund fish included uncoordinated spiral swimming, violent flexing of the body and development of anaemia. Diseased fsh were notably darker in colour and appeared weak and lethargic. Acute infection was systemic, and the septic process developed caused lesions in almost all organs and tissues. Liver, kidney and digestive tract lesions were noted. Clinical symptoms of disease included muscle lesions, gill oedema, internal organ deformation, and tissue necrosis. The cumulative mortality in inspected fshfarms was 60%–70%.
The reproduction of isolated virus in continuous cultures of RTG-2, FHM and EPC fsh cells was investigated. All three cell lines were sensitive to the virus which caused morphological changes, such as vacuole enlargement and cell rounding. Subsequently, cells detached from the surface and characteristic CPE of virus on cells was visible. For cell lines of RTG-2 and FHM the complete destruction of monolayer was noted 7–8 days after infection (d.a.i.). In EPC cultures, characteristic CPE and complete destruction of cell monolayer were noted 10–12 d.a.i. (Figure 1A). The infectious titre of IPNV "Karpaty" was 105.5–5.8 TCID50/mL for EPC, and 106.2–6.5 and 106.9–7.4 TCID50/mL, respectively, for the FHM and RTG-2 cell lines. The low infectious titre of virus in EPC cells can be related to its slow reproduction in this culture. The greatest infectious titre was observed for the culture of RTG-2, very appropriately, as this cell line was derived from a rainbow trout – the natural IPNV reservoir. That is why RTG-2 is the most appropriate cell line for accumulation of the Ukrainian isolate IPNV "Karpaty". However, all three cell cultures of RTG-2, FHM and EPC can be used for diagnosis of the Ukrainian IPNV isolates.
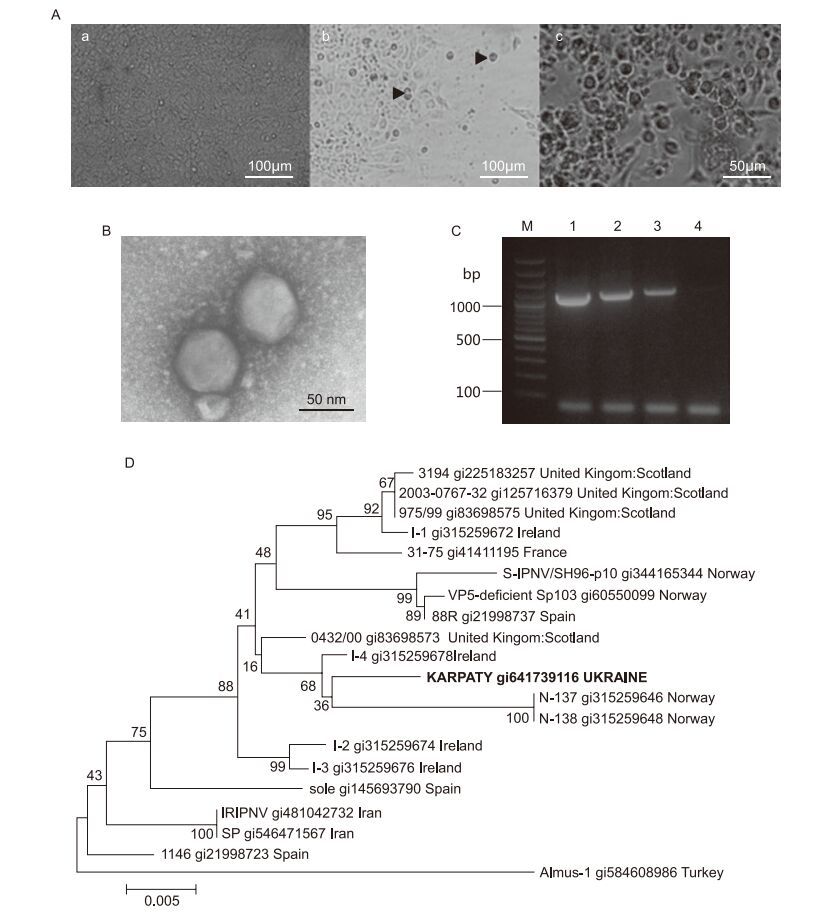
Figure 1. (A) Changes in EPC cell monolayer during IPNV infection. Non-infected cells (a), 5 d.a.i. (b) and 10 d.a.i. (c) with Ukrainian isolate IPNV "Karpaty": arrows indicate cell rounding and detachment from a surface. Virus titre at 10 d.a.i. was 105.5–5.8 TCID50/mL (×200). (B) Electron microscopy of particles of IPNV strain "Karpaty" after virus purifcation (×80000). Negative staining with uranyl acetate. (C) Amplifcation of IPNV genomic RNA by RTPCR: lanes 1–3: Ukrainian isolate IPNV "Karpaty", predicted size of amplifed VP2 gene fragment=1120 bp; lane 4: no template, negative control; lane M: GeneRuler 100 bp Plus DNA ladder (ThermoScientifc). (D) Phylogenetic analysis of VP2 gene of IPNV isolate "Karpaty". The tree was generated by means of the neighbour-joining algorithm in MEGA software version 6.0.
Results of our electron-microscopy investigations of purifed viral particles revealed morphology and ultrastructure characteristics typical of birnaviruses. Virions of the Ukrainian isolate of IPNV "Karpaty" were non-enveloped, icosahedral, and approximately 70 ± 5 nm in diameter (Figure 1B).
PCR was used for identification of isolated birnavirus. The primer set targeting the full length VP2 gene was selected for amplification of viral genomic dsRNA. Amplifed fragments were approximately 1120 bp in size (Figure 1C).
The nucleotide sequences of amplifed fragments were analysed and the identity of the Ukrainian-isolated IPNV to the Sp strain was revealed. The nucleotide sequence was registered in GenBank under accession number KJ596654. The virus was named IPNV isolate "Karpaty". Comparison of sequences of IPNV VP2 genes from NCBI with amplifed fragments of IPNV isolate "Karpaty" confrmed the high identity of 99% with the Sp strain. Among the isolates of the Sp strain, the most closely related to IPNV "Karpaty" were viruses found in Norway, Ireland and the United Kingdom (Figure 1D).
In our opinion, for rapid virus identification by PCR, WB primers (Williams et al., 1999) should be used, because in our previous research (unpublished data) only WB primers were able to identify the virus at low concentrations of target RNA. Further monitoring of IPNV and its diagnosis in salmonids cultivated in fsh-farms or native ponds in the Ukraine are required in order to accumulate data concerning virus distribution and also to identify other strains widespread in Europe. It will be the subject of our future research.
HTML
-
This study was partially supported by the Fisheries Agency of the Ukraine. We thank Ms. Antonina I. Mruk for help in sample collection. The authors declared that they have no conflict of interest. This article does not contain any study with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: