HTML
-
Foamy viruses (FVs) are complex retroviruses and the only genus of the Spumaretrovirinae subfamily (Linial, 1999). FVs have a wide host range and are endemic in most non-human primates (Broussard et al., 1997; Saib, 2003), cats (Mochizuki et al., 1990), cattle (Johnson et al., 1983), and horses (Tobaly-Tapiero et al., 2000). FVs have two promoters: a conserved promoter in the 5' long terminal repeat (LTR) that is similar to that found in other retroviruses and a unique internal promoter (IP) (Lochelt et al., 1994). The IP controls the transcription of the regulatory proteins Tas and Bet during the early stage of viral transcription. The FV Tas protein is a DNA-binding protein that binds specifically to the transactivation responsive elements (TREs) in the 5' LTR and IP. Thus, FV Tas can engage in a positive feedback loop through the IP to enhance its own expression and increase expression of other structural proteins (Rethwilm, 2010).
Bovine foamy virus (BFV) is a member of the Spuma-virus genus that was first isolated from cattle in 1983. BFV encodes three structural genes (gag, pol, and env) and two regulatory genes (btas and bbet). Viral gene expression is regulated primarily by BTas, the transactivator protein. BTas is a 249-amino acid (aa) protein that contains two domains: the N-terminal DNA binding domain (1–133 aa) and the C-terminal activation domain (198–249 aa) (Tan et al., 2010). The transactivator activity of BTas is dependent on both its ability to bind DNA and its ability to dimerize (Tan et al., 2008a; Tan et al., 2010).
Retroviral proteins have recently been identified as targets for regulation by cellular histone acetyltransferase. For example, three lysine residues (positions 264, 266, and 273) in the HIV integrase protein are acetylated by p300, which results in increased DNA binding and promotes DNA strand transfer activity (Cereseto et al., 2005). Similarly, acetylation of the HTLV-1 Tax protein enhances signaling through the NF-κB pathway (Lodewick et al., 2009). The coactivators p300 and PCAF also acetylate Tas, leading to enhanced Tas-mediated activation in prototype foamy virus (PFV) infection (Bannert et al., 2004), and PCAF plays a role in acetylating the feline foamy virus (FFV) Tas in vitro and in vivo, which enhances the ability of Tas to bind to the viral promoters (Bodem et al., 2007). Finally, our group previously demonstrated that BTas is acetylated by p300 in vivo and in vitro (Chang et al., 2011). Acetylation is required for BTas to interact with DNA, and overexpression of p300 enhanced BTas-mediated activation of the BFV LTR and IP. Our laboratory also demonstrated that p300-mediated acetylation of BTas occurs at three lysine residues in positions 66, 109, and 110 (Chang et al., 2011). However, the importance of BTas acetylation to BFV replication has not been well studied. Therefore, the aim of this study was to establish whether the lysine targets for acetylation in BTas are important for the transactivation function of BTas and viral replication.
-
The BFV indicator cell line (BFVL) is a modified BHK-21 cell line that contains a luciferase reporter gene under the control of the BFV LTR promoter (Guo et al., 2011). BICL, a second reporter cell line also based on the BHK-21 cell line, contains an EGFP gene integrated into the genomic DNA driven by the BFV LTR (Ma et al., 2007). HEK 293T, BHK-21, BICL, and BFVL cells were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco, Gr and Island, USA) supplemented with 10% (v/v) fetal bovine serum, 50 IU/mL penicillin, and 50 μg/mL streptomycin at 37 ℃ in a 5% CO2 atmosphere. HEK 293T, BHK-21, and BICL cells were transfected using polyethylenimine (PEI; Polysciences, Warrington, USA) per the manufacturer's protocol.
-
A single infectious BFV clone containing the BFV3026 proviral genome in pBluescript SK-(pBS-BFV) was used as the starting point for all mutations (Bing et al., 2014). An Nde I restriction site was used to divide the full-length BFV genome into two segments, S1 and S2, and mutations were introduced in the S2 segment. To strea-mline the polymerase chain reaction (PCR) -based site-directed mutagenesis, S2 was subcloned into the smaller pMD18T vector. PCR-based site-directed mutagenesis was used to introduce two stop codons using the primers ΔBTas-upper and ΔBTas-lower (Table 1). The desired mutations were confirmed by DNA sequencing. The mutated S2 fragment was then used to replace the original S2 fragment in the full-length infectious clone pBS-BFV. The final BTas-deficient clone was termed pBS-BFV-ΔBTas. For "add back" assays to recover BTas func-tion when cells were transfected with pBS-BFV-ΔBTas, wild-type BTas was expressed from the plasmid pcDNA3.1-BTas (Tan et al., 2008a). pcDNA3.1-BTas and BFV clones containing the lysine-to-arginine (K→R) mutations that were previously shown to inhibit acetylation of BTas (K66R, K109R, K110R, and K66/109/110R [also called 3M]) were generated using PCR-based site directed mutagenesis (Chang et al., 2011). The primers used for the K→R mutations are shown in Table 1. DNA sequencing was used to confirm that the desired sequence was achieved for all constructs.
Primers Sequence (5′-3′) ΔBTas upper GTAAGGAAGACCTCATAT A AT AACTTGCTTGTACCAΔBTas lower TGGTACAAGCAAGTT A TT ATATGAGGTCTTCCTTACK66R upper GAATTATAAACTTCATCTCA G ACGAGATGAACTTAGAGGAGK66R lower CTCCTCTAAGTTCATCTCGT C TGAGATGAAGTTTATAATTCK109R upper GTCATTAGAAACA G AAAGGGGCCATACK109R lower GTATGGCCCCTTT C TGTTTCTAATGACK110R upper CATTAGAAACAAAA G GGGGCCATACATGK110R lower CATGTATGGCCCC C TTTTGTTTCTAATGK109/110R upper GTCATTAGAAACA G AAG GGGGCCATACK109/110R lower GTATGGCCCC C TTC TGTTTCTAATGACNote: The mutant nucleotides are indicated with boxes. The underlined nucleotides indicate the positions of the mutated codons. Table 1. Primers used in this study
-
Cell lysates were separated by SDS-PAGE. The separated proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane (GE, Freiburg, Germany) by electroblotting for 1 h at 100 V. The PVDF membranes were blocked in 5% non-fat milk-PBS for 45 min at room temperature and subsequently incubated with primary antibody for 90 min. The primary antibodies against BTas (Tan et al., 2008b), Bet, and Gag (Wang et al., 2010) were generated in our lab and have been described previously. The secondary antibody was either a goat anti-mouse or goat anti-rabbit IgG conjugated to peroxidase. The membranes were incubated with secondary antibodies for an additional 45 min. Protein signals were detected by chemiluminescence (Millipore, Billerica, USA).
-
The infectivity of each BFV infectious clones was analyzed using two different BFV indicator cell lines. BICL cells were transfected with pBS-BFV infectious clones, and the GFP signals were detected at different times post-transfection. BHK-21 cells or BICL cells were transfected with pBS-BFV plasmids. The transfected cells were harvested and then co-cultured with the BFVL reporter line. After 48 h co-cultivation, the co-cultured cells were harvested and subjected to luciferase assays according to the manufacturer's protocol (Promega, Madison, USA). The luciferase activity was normalized to the uninfected value. At least three independent experiments were performed, and the data were presented as means ± standard deviations.
-
HEK 293T cells were transfected with either wild-type or mutant BTas plasmid and either the LTR-Luc or IP-Luc plasmid, which expressed firefly luciferase under the control of the BFV LTR or IP promoter, respectively. Transfection efficiency was normalized to the expression of the pCMV-β -gal plasmid, which expresses β -Gal from the CMV promoter and was co-transfected with the BTas-and luciferase-expressing plasmids. Luciferase activity was measured 48 h post-transfection and normalized to β -gal activity.
-
BICL cells were plated at a density of approximately 2.0–2.5×105 per well in six-well plates in DMEM medium. Twenty-four hours later, the cells were transfected with pBS, pBS-BFV, or infectious clones containing the mutant BTas genes. BICL cells were passaged when they reached approximately 95% confluency. The GFP signal in the cells was assessed by fluorescence microscopy (Olympus Ⅸ-71, Tokyo, Japan) at 2, 4, and 6 days post-transfection to detect BFV transactivation. BICL cells transfected for 2, 4, and 6 days were also collected and co-cultured with BFVL cells.
Cell culture, virus, and transfection
Construction of mutated BFV molecular clones
Western blotting
Measuring BFV infectivity
BTas transactivation assays
Virus passage
-
It is well established that Tas is indispensable for PFV (Lochelt et al., 1991) replication. We previously showed that BTas could transactivate BFV transcription (Liu et al., 1999), suggesting that BTas is required for BFV replication. However, in the studies of BFV infectious clones, there is no direct evidence that BTas is required for viral replication. Therefore, a BTas-deficient BFV molecular clone, pBS-BFV-ΔBTas was generated, by inserting two stop codons after codon 42 of the BTas gene, leading to the expression of a non-functional short peptide (Figure 1A). The mutation did not change the expression of other viral genes or affect the splicing of viral RNAs (data not shown). Then the BTas-deficient clone was transfected into HEK 293T cells and the viral protein expression was assessed by western blotting at 48 h post-transfection. As expected, the BTas protein was not detected in HEK 293T cells transfected with pBS-BFV-ΔBTas, nor were the BFV Bet and Gag proteins (Figure 1B). To confirm these findings, wild-type or BTas-deficient pBS-BFV were transfected into BHK-21 cells (BFV-permissive cell line). BHK-21 cells were harvested 48 h post-transfection and subsequently co-cultured with the BFVL reporter cell line. A luciferase assay was performed after a 48-h co-cultivation to determine BFV infectivity. As shown in Figure 1C, BTas-deficient viruses were unable to replicate.
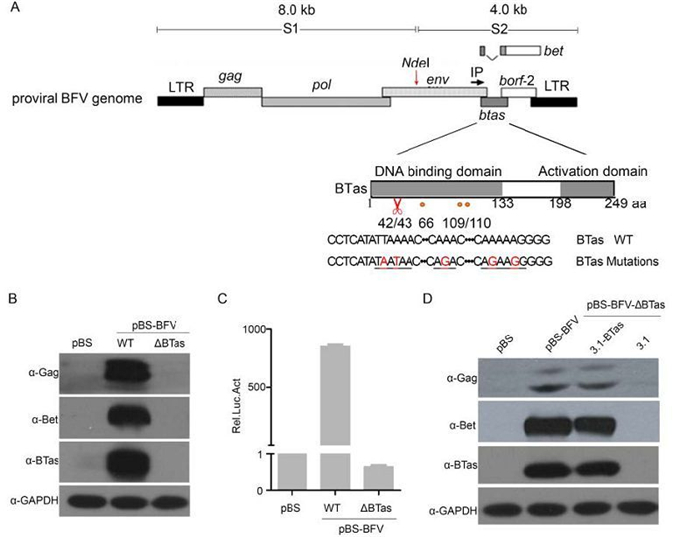
Figure 1. Deletion of BTas from full-length infectious BFV abolished viral replication. (A) Schematic representation of the BFV3026 proviral genome and the mutation sites within the btas gene (indicated in red). The red scissors indicate the position where the premature stop codon was inserted. (B) HEK 293T cells were transfected with the empty vector (pBS) or BFV infectious clones that were wild type (WT) or BTas deficient (∆BTas). Viral protein expression was evaluated by western blotting 48 h post-transfection. (C) BHK-21 cells were transfected with BFV infectious clones (WT or ΔBTas) and then co-cultured with BFVL reporter cells 48 h post-transfection. The luciferase activity in the co-culture was normalized to that of the empty vector. The relative luciferase activity (Rel. Luc. Act) is shown for each condition. Data are presented as the mean ± standard deviation. At least three independent experiments were performed. (D) HEK 293T cells were co-transfected with a BTas-deficient BFV clone (pBS-BFV-ΔBTas) and either empty vector (pcDNA3.1) or a wild-type BTas expression plasmid (pcDNA3.1-BTas). After 48 h, the transfected cells were lysed, and viral protein expression was assessed by western blotting.
An "add back" experiment was performed to confirm that the loss of BTas was directly responsible for the loss of viral protein expression. HEK 293T cells were co-transfected with the BTas-deficient clone (pBS-BFV-ΔBTas) and a plasmid encoding BTas (pcDNA3.1-BTas). When the pcDNA3.1-BTas plasmid was co-transfected with the BTas-deficient BFV clone, expression of the Gag and Bet proteins was detectable (Figure 1D, lanes 3 and 4). Taken together, these results demonstrate that BTas-deficient viruses have a replication defect and suggest that BTas is essential for BFV replication.
-
We previously demonstrated that p300 acetylates lysine residues in BTas at positions K66, K109, and K110 and that acetylation of these residues enhances BTas DNA-binding affinity. In this study, to address whether these residues are critical for the transactivation function of BTas, we generated mutant proteins that substitute arginine for lysine (K→R) at each individual position and a triple mutant that replaces all three lysine residues with arginine. Arginine was chosen for the mutants because it is a positively charged amino acid similar to lysine. HEK 293T cells were co-transfected with wild-type or mutant BTas plasmids and either the BFV LTR-Luc or IP-Luc plasmid. When a single lysine residue was mutated, activation of the BFV LTR-Luc plasmid was reduced approximately 40%–50% compared to wild-type BTas (Figure 2A). The triple mutant with all three lysine residues mutated to arginine lost approximately 80% of the luciferase reporter activity. The impact of replacing a single lysine residue on luciferase activity from the IP was slightly less than for the LTR: approximately a 30% reduction compared to wild-type BTas. However, the triple mutant reduced the luciferase reporter expression from the IP-Luc construct approximately 70% (Figure 2B). The expression levels of the wild-type and mutant BTas proteins were similar (Figure 2). These results indicate that the lysine residues at positions 66, 109, and 110 are critical for BTas transactivation.
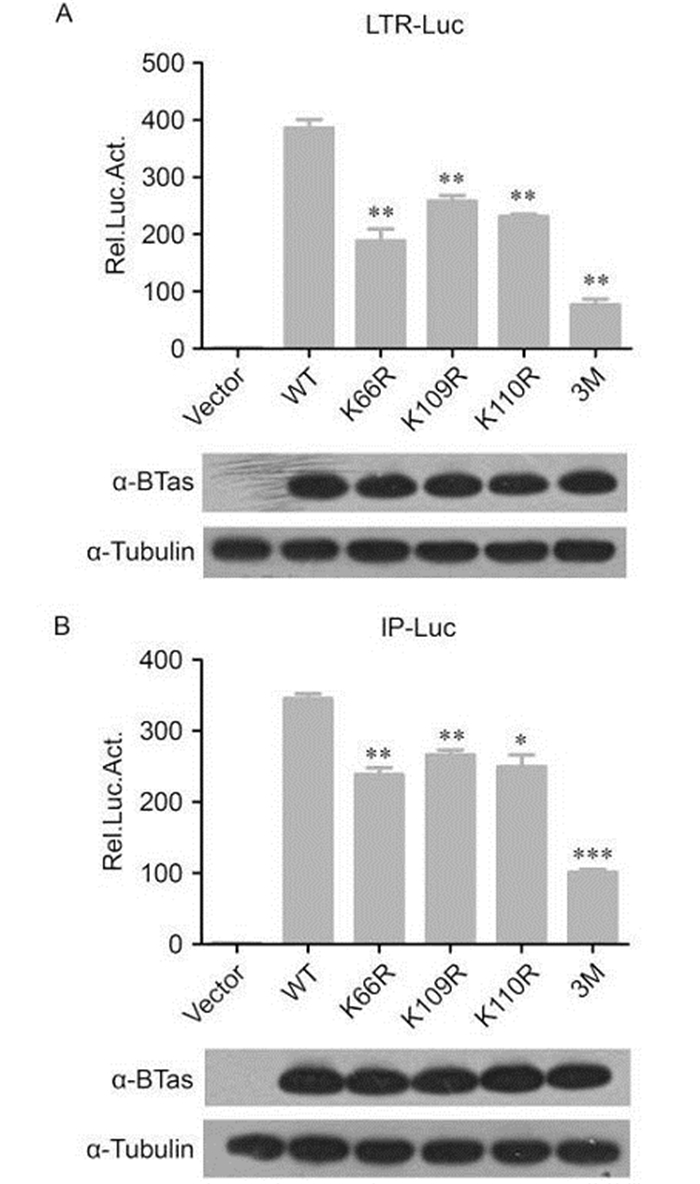
Figure 2. BTas-dependent transactivation requires lysine residues at positions 66, 109, and 110. HEK 293T cells were co-transfected with a plasmid expressing wild-type (WT) BTas or point mutants containing lysine to arginine (K→R) mutations at position 66 (K66R), 109 (K109R), 110 (K110R), or all three positions (3M) and a luciferase reporter under the control of the conserved 5' LTR promoter (A) or the unique BFV IP promoter (B). β-gal expression from the pCMV-β-Gal vector was used as an internal control for transfection efficiency, and the luciferase expression was normalized to β-gal. Luciferase activity was measured 48 h post-transfection. Data are presented as the mean ± standard deviation. At least three independent experiments were performed. Significant differences between the WT virus and the mutants were identified using Student's t-test. The threshold for significance was established at P < 0.05. *P < 0.05, **P < 0.01, ***P < 0.001.
-
To determine whether K66, K109, and K110 are important for BFV replication, we generated single K→R mutants at position 66, 109, or 110 and a triple K→R mutant (3M) in the pBS-BFV infectious clone. BICL cells were transfected with the wild-type pBS-BFV clone and each of the mutants. The BICL cells were harvested after 48 h and then co-cultured with BFVL cells to determine viral infectivity. Viral protein expression was also assessed in BICL cells by western blotting. The wild-type and mutant viruses expressed similar levels of BTas in BICL cells 48 h post-transfection. However, the expression level of Gag decreased in all of the mutant BFV clones (Figure 3A). The infectivity of the single K→R mutant BFVs was also impaired: the K66R, K109R, and K110R mutants were approximately 20%, 50%, and 75% as infectious as wild type, respectively. The 3M mutant lost most of its ability to replicate (Figure 3B).
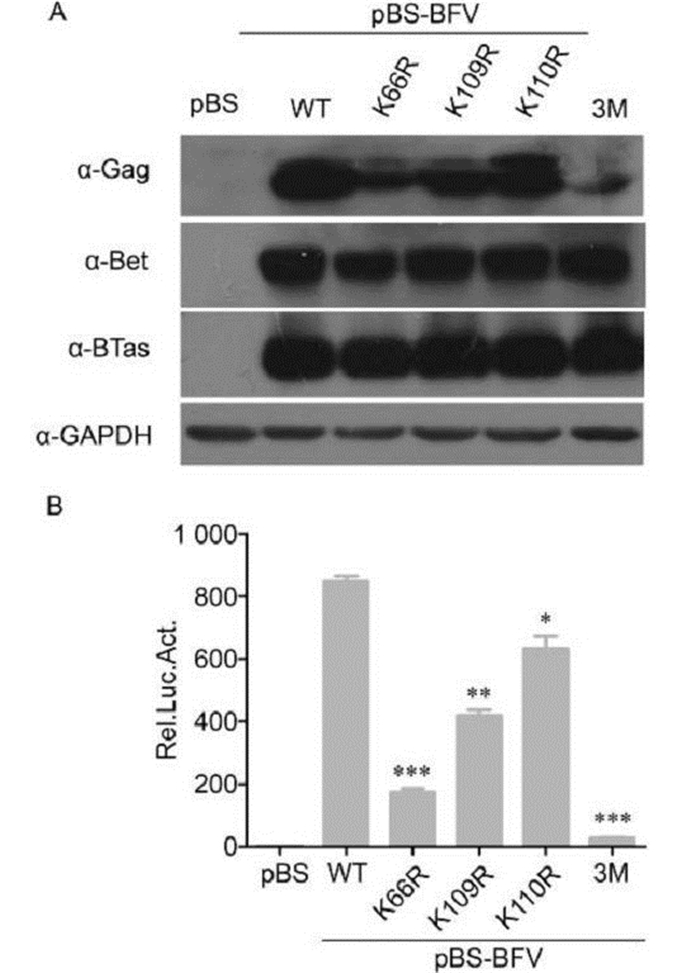
Figure 3. Lysine residues at positions 66, 109, and 110 in BTas were required for BFV replication. (A) BHK-21 cells were transfected with empty vector or BFV infectious clones that were wild-type (WT) or lysine to arginine (K→R) point mutations at position 66 (K66R), 109 (K109R), 110 (K110R), or all three (3M). After 48 h, viral protein expression was evaluated by western blotting. (B) Transfected BHK-21 cells were harvested and co-cultured with BFVL cells. Luciferase activities were measured at 48 h post-infection. Data are presented as the mean ± standard deviation. At least three independent experiments were performed. Significant differences between the WT virus and the mutants were identified using Student's t-test. The threshold for significance was established at P < 0.05. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, whether the K→R mutations would regain infectivity as high as wild type during long-term virus passage was assessed. BICL cells were transfected with wild-type or mutant BFV clones, and the degree of BFV replication was assessed by measuring GFP expression at 2, 4, and 6 days post-transfection. Both the K66R and 3M mutants showed reduced replication activity compared to wild-type BFV at 2 days post-transfection, and there was no restoration in the subsequent passage (Figure 4A). The level of infectious virus production was also determined by co-culturing transfected BICL cells with BFVL cells. The degree of BFV infection in the BFVL cells was measured by luciferase assay. The results were consistent with those from the fluorescence experiments (Figure 4B). Together, these data suggest that lysine residues at positions 66, 109, and 110 of BTas are indispensable for BFV replication.
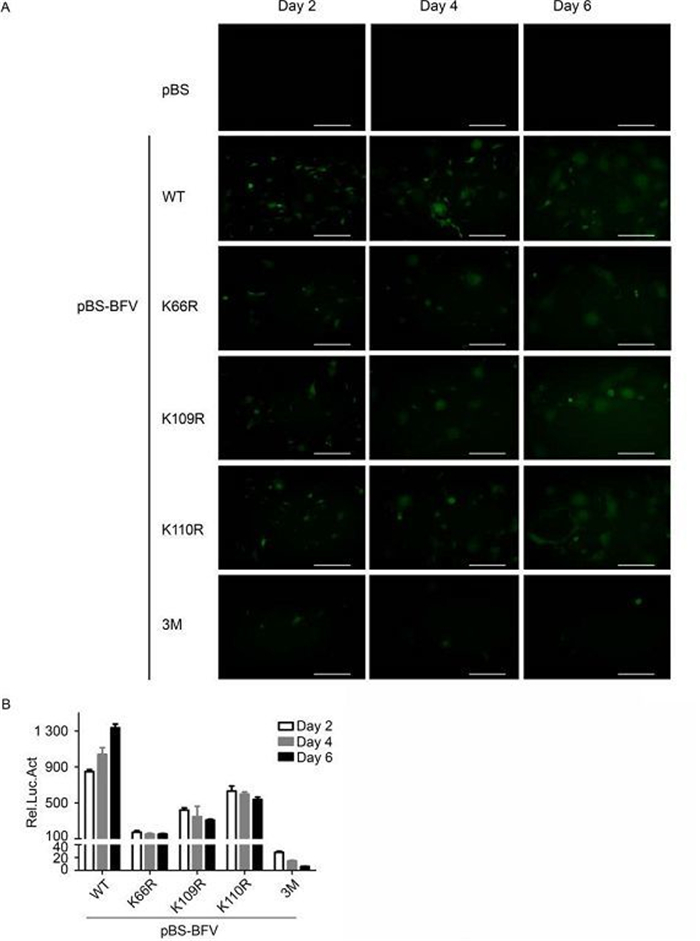
Figure 4. Lysine residues at positions 66, 109, and 110 in BT as affect the passage of BFV. (A) BICL cells were transfected with empty vector, or BFV infectious clones that were wild-type (WT) or lysine to arginine (K→R) point mutants. Viruses were kept in culture for 6 days. Green fluorescence was detected at days 2, 4, and 6 post-transfection. (B) BICL cells were harvested and co-cultured with BFVL cells. Virus infection in the BFVL cells was determined by luciferase assay. Data are presented as the mean ± standard deviation. At least three independent experiments were performed. Scale bars indicate 300μm.
Deletion of BTas in full-length infectious clones abolished BFV replication capacity
BTas-dependent transactivation requires lysines at positions 66, 109, and 110
Lysine residues at positions 66, 109, and 110 in BTas are important for BFV replication
-
The importance of BTas as a central regulatory protein that contributes to the transactivation of the BFV LTR and IP has been well described (Liu et al., 2000; Zhang et al., 2000). Herein, we provide direct evidence that BTas is critical to BFV replication using a prematurely truncated form of BTas. Viral replication was restored when wild-type BTas was supplied on a co-transfected plasmid, indicating that the replication defect observed with the BTas-deficient mutant BFV clone was attributable to the loss of BTas. The BTas used in this paper has higher transactivation ability (unpublished data) than the protein used in our previously published work (Chang et al., 2011). This may explain why there is still modest transactivation ability in the 3M BTas mutant, whereas our previous study showed a complete loss of this activity.
We previously showed that p300 and BTas act synergistically to activate the BFV promoters and that p300 acetylates lysine residues at positions 66, 109, and 110 in BTas, which is required for DNA binding. In addition, the lysine residue at position 110 is thought to be particularly important, as K110 mutants alone or in combination with K66 and K109 mutations abrogate the ability of BTas to bind DNA in vitro (Chang et al., 2011). In this study, we show that K→R substitutions at each of K66, K109, and K110 or as a triple mutant reduce the ability of BFV to transactivate gene expression from the LTR and IP in vivo. However, in this study, the K66 mutation has the greatest impact. Similar results were obtained for BFV replication. The apparent discrepancies between the in vitro and in vivo data suggest that K66 might have additional unknown functions in the BFV life cycle. Our findings also suggest that acetylation might be contributing to BTas function. The K→R mutation prevents acetylation at K66, K109, and K110; although we cannot rule out structural changes arising from the K→R mutation that affect BTas function, the in vitro data suggest that the loss of BTas acetylation contributes to the reduced DNA binding activity and reduced BFV replication. This interpretation would be consistent with findings from the HIV-1 field indicating that acetylation of different lysine residues in the HIV-1 Tat protein regulates different functions (Col et al., 2001; He et al., 2013) and with Tas and Tat-mediated transcription in PFV and HIV-1, respectively (Ott et al., 1999; Bannert et al., 2004).
In summary, our work is the first to show that BTas is an essential factor for BFV replication using BTas deficient molecular BFV clone. We also demonstrated that BTas-dependent transactivation and BFV replication require acetylation lysines at BTas positions 66, 109, and 110. Together with our previous study showing that BTas is acetylated at lysines 66, 109, and 110, this study suggest that acetylation is a ubiquitous mechanism adopted by FVs to facilitate viral replication.
-
This work was supported by grants from the National Natural Science Foundation of China (Na. 31370182), Tianjin Research Program and Application Foundation and Advanced Technology (Na. 12YFQNJC02300), and 111 Project (B08011).
-
All the authors declare that they have no competing interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
SZZ, XXC and JL participated in the experiments and data analysis. SZZ and ZBL drafted manuscript. WTQ and JT conceived of the study. All authors read and approved the final manuscript.














 DownLoad:
DownLoad: