HTML
-
Human immunodeficiency virus type 1(HIV-1)can rapidly generate mutants and evade immune responses(Vider-Shalit et al., 2009). This is the major obstacle to the development of prophylactic HIV-1 vaccines. Design of HIV-1 vaccine immunogens that can elicit high-titer, potent, broadly neutralizing antibodies(bnAbs)that inhibit viruses from different genetic subtypes is widely agreed(Liao et al., 2006; Zhang et al., 2008). Most of bnAbs primarily bind to membrane proximal external region(MPER)of glycoprotein 41(gp41), glycans or CD4 binding site(Eroshkin et al., 2014). Several identified bnAbs, such as b12(Roben et al., 1994), X5(Moulard et al., 2002), 2G12(Trkola et al., 1996), 2F5 and 4E10(Stiegler et al., 2001; Zwick et al., 2001), exhibit potent, broad HIV-1 neutralizing activity in vitro and can prevent HIV-1 infection in animal models. These bnAbs target HIV-1 envelope glycoproteins(Envs)that are crucial for virus-cell fusion. Therefore, various Envs are potential c and idate immunogens and have been evaluated in animal models and human clinical trials(Trkola et al., 1996; Stiegler et al., 2001; Zwick et al., 2001; Moulard et al., 2002). However, failure to elicit bnAbs by using these Envs as immunogens reflects the difficulty to produce a vaccine that is effective in humans(Mehandru et al., 2004). It has been suggested that extensive characterization of bnAb-binding epitopes will help in the design of effective vaccine immunogens that are able to elicit bnAbs or similar antibodies in vivo(Alam et al., 2007). Although this approach is being pursued vigorously, few HIV-1 immunogens have efficiently produced neutralizing antibodies with broad specificity. For example, no vaccine or immunization strategy has been found to induce robust bnAb responses against gp41 MPER. A major aim of HIV-1 vaccine research is to determine how such rare bnAb responses are elicited.
The gp41 is a subunit of the HIV-1 Env complex and plays an important role in cell entry. The MPER is one of the most highly conserved sequences of gp41 that is crucial to the fusion of the viral and cellular membranes. MPER is a target for the neutralizing monoclonal antibodies 2F5, 4E10, Z13 and m66.6, making it a considerably interested epitope for HIV-1 vaccine development. The 2F5 epitope localizes on MPER spreading from amino acids(aa)662 to 667(sequence ELDKWA)(Montero et al., 2012), and that of m66.6 spans the 2F5 epitope and two additional upstream residues(L660 and L663)(Zhu et al., 2011). However, m66.6 exhibits less potency and narrower specificity than 2F5 bnAb. The existence of bnAbs such as 2F5 has fueled the hope that the development of efficacious HIV vaccines is achievable using immunogens comprising the epitopes recognized by these bnAbs(Chen et al., 2010). Unfortunately, failure so far to induce a high-titer neutralizing response in vaccination with antigenic envelope constructs expressing the 2F5 epitope reflects a lack of underst and ing of the mechanism of 2F5-like antibody evolution(Prabakaran et al., 2007; Shen et al., 2009). There is only sparse information about specific patients from whom HIV-1 bnAb specificities have been obtained. Therefore, the finding of novel bnAbs and their conserved epitopes still has implications for the design of HIV-1 vaccine immunogens and is also useful for underst and ing the mechanisms of HIV entry into host cells and evasion of immune responses.
In this study, we describe the identification and characterization of two human 2F5-like antibodies against the MPER antigen in yeast display antibody libraries constructed from a HIV-infected subject. The library was screened against peptide SP62 containing the 2F5 epitope. The identified 2F5-like antibodies weakly neutralized entry of virions pseudotyped with Envs from HIV-1 primary isolates. We speculate that the presence of residues aa652-655 adjacent to the 2F5 epitope influences the immune response of eliciting neutralizing antibodies like 2F5. These data will be useful in the design of MPER specific vaccine immunogens capable of eliciting bnAbs in vivo.
-
Escherichia coli(E. coli)TOP10 was used for cloning and preparation of plasmid DNA. E. coli HB2151 was used for expression of soluble single-chain fragment variable antibody(scFv) and single chain Fab antibody(scFab). HEK293T was purchased from ATCC. Other cell lines and plasmids for expression of various HIV-1 Envs were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program(ARRRP). Three biotinylated peptides, SP62(QQEKNEQELLELDKWASLWN), SCR(the amino acid sequence of SP62 scrambled), and MPER656(NEQELLELDKWASLWNWFNITNWLW), were synthesized by Genscript. Horseradish peroxidase(HRP)-conjugated anti-FLAG antibody and HRP-conjugated anti-human IgG(Fc-specific)antibody were purchased from Sigma-Aldrich. Alexa-488 goat anti-mouse IgG antibody, mouse anti-c-Myc antibody and streptavidin R-phycoerythrin conjugate were obtained from Invitrogen.
-
cDNA was reverse transcribed from total RNA of peripheral blood monouclear cells isolated from a HIV-1 infected patient. Heavy-and light-chain of antibody genes were PCR amplified from cDNA using a primer set based on the protocol of Zhu and Dimitrov(Zhu and Dimitrov, 2009). The construction of scFv and scFab genes was as described previously(Walker et al., 2009). The preparation of yeast libraries was performed according to our previous publications(Zhao et al., 2012; Zhao et al., 2015). The size of the scFv and scFab libraries was estimated at 4 × 106 and 1 × 106 members, respectively.
-
Construction and culture of yeast libraries for affinity maturation were as described previously with some modifications(Zhao et al., 2011; Zhao et al., 2012). Briefly, yeast cells were grown in SD-CAA media, and induced in SG-CAA in volumes appropriate for the size of the library(Chao et al., 2006). For library selection, induced yeast cells(1 × 109)were incubated in 3 mL of PBSA buffer(0.1% BSA in PBS)with 1 μg/mL peptide SP62 for 1 h at 4 ℃ with gentle rotation. Excess peptides were washed away in cold PBSA, and then cells were mixed with 50 µL Dynabeads MyOne Streptavidin T1 beads(Invitrogen)in 3 mL PBSA buffer. Capture was performed at 4 ℃ for 1 h with gentle rotation. A magnet was then applied to select out the beads, and the supernatants were removed. After washing 3 times with ice-cold PBSA buffer, the cell pellet was cultured in 10 mL SD-CAA media at 30 ℃ overnight. The yeast cells recovered from the magnetic bead were induced in 50 mL SG-CAA for 16-18 h at 20 ℃. The following two to three rounds of sorting were performed by repeating the above steps. After the final round of selection, yeast cells were cultured on SD-CAA plates and individual colonies were picked for characterization.
Binding of yeast cells was analyzed using flow cytometry. Typically, 1 × 106 cells were stained in 50 µL of PBS in the presence of 0.1 µg/mL biotinylated peptide and 5 µg/mL mouse anti-c-Myc antibody with 30 min incubation and washed with 200 µL PBSA buffer. For competition analysis, 2F5 IgG was pre-incubated with peptides and then incubated with cells for 30 min at 4 ℃. Yeast cells were washed and then incubated with 5 µg/mL Alexa-488 goat anti-mouse IgG antibody(Invitrogen) and 10 µg/mL streptavidin R-phycoerythrin conjugate. The cells were resuspended in PBSA buffer and counted using a BD FACSCalibuTM Flow Cytometer.
-
Isolation of plasmid DNA from individual yeast cells was performed using the Zymoprep Yeast Plasmid Miniprep Kit Ⅱ(Zymo Research). The plasmids were then transformed into competent E.coli TOP10 cells for amplification of plasmid DNA which was purified using the QIAprep Spin Miniprep Kit(Qiagen). Individual DNA inserts were obtained from plasmids digested with the restriction enzyme Sfi Ⅰ and ligated into similarly digested vector pComb3X for soluble expression in E. coli HB2151. scFv and scFab were expressed and purified as previously described(Zhao et al., 2009). The supernatant was used for purification of scFv and scFab by immobilized metal ion affinity chromatography(IMAC)using Ni-NTA resin(Qiagen)according to the manufacturer's protocols.
Cell lines, peptides, and antibodies
Yeast library construction
Library selection and flow cytometric analysis
Expression and purification of antibodies
-
Various concentrations of antigens were diluted in PBS buffer and coated onto 96-well ELISA plates at 4 ℃ overnight. The plate was then blocked with PBS containing 3% powdered milk buffer. Antibodies were serially diluted in the same blocking buffer and applied to the 96-well ELISA plates. Mouse anti-Flag-HRP was used to detect Flag tag at the C-terminal end of each of the scFvs or scFabs, and mouse anti-human Fc-HRP was used to detect Flag tag. The 2, 2'-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt(ABTS)solution was then added to each well and optical density at 450 nm(OD450)was measured 10 min later.
-
Viruses pseudotyped with HIV-1 Envs were prepared by cotransfection of 70%-80% confluent 293T cells with pNL4–3.luc.E-R- and pSV7d constructs encoding HIV-1 Envs by using the PolyFect transfection reagent(Qiagen), according to the manufacturer's instructions. Pseudotyped viruses were obtained after 48 h by centrifugation and filtration of cell culture through 0.45-μm filters. For neutralization assays, viruses were mixed with different concentrations of antibodies for 1 h at 37 ℃, and then the mixture was added to about 1.5 × 104 HOS-CD4-CCR5(used for all R5 and dual tropic viruses)cells grown in each well of 96-well plates. Luminescence was measured after 48 h using the Bright-Glo Luciferase Assay System(Promega) and a LumiCount microplate luminometer(Turner Designs). Mean relative light units(RLU)were determined for duplicate wells. Relative infectivity(%)was calculated by the following formula:(average RLU of antibody-containing wells/average RLU of virus-only wells)× 100.
Pseudovirus neutralization assay
-
The plasmid used for library construction, pYD7, was modified from pCTCON2(Hackel et al., 2008). Two Sfi I restriction sites at both ends of the scFv DNA fragment in pYD7 matched the cloning sites in the pComb3X vector. This allowed fragments to be shuttled between the yeast vector and pComb3X. The linkers(G4S)3 and (SGGG)2(SEGGG)4(SGGGSG)(Walker et al., 2009)were inserted between the heavy and light chains of antibody, generating scFv and scFab respectively. The scFv(4 × 106) and scFab(1 × 106)yeast libraries were constructed from peripheral blood B cells of an HIV-1 infected patient who exhibits broad serum neutralization. To estimate the expression of libraries, library yeast cells were stained with mouse anti-c-Myc antibody and then counted using flow cytometry. More than 40% of the yeast clones were expressed in the scFv library, but only around 10% of the yeast clones were expressed in the scFab library.
-
The scFv and scFab libraries were subjected to multiple rounds of magnetic bead selection against MPER peptide SP62 according to the protocol of Yeung and Wittrup(Yeung and Wittrup, 2002). Significant enrichment was obtained from the scFv library after three rounds selection(Figure 1A), but enrichment was not obvious from the scFab library after four rounds selection(Figure 1B). Following the final round of selection, polyclonal yeast cells were cultured for characterization. The 2F5 IgG antibody was used to determine whether enriched clones bound the 2F5 epitope on MPER peptide SP62. Eight of 36 clones from the scFv displayed library were identified to bind to the SP62 peptide. Sequence analysis of these eight clones showed that they represented two individual clones, 3A2a and 3A5a(Figure 2A, 2B), containing identical light chains of the IGKV1 family and very closely related heavy chains of the IGHV3 family.
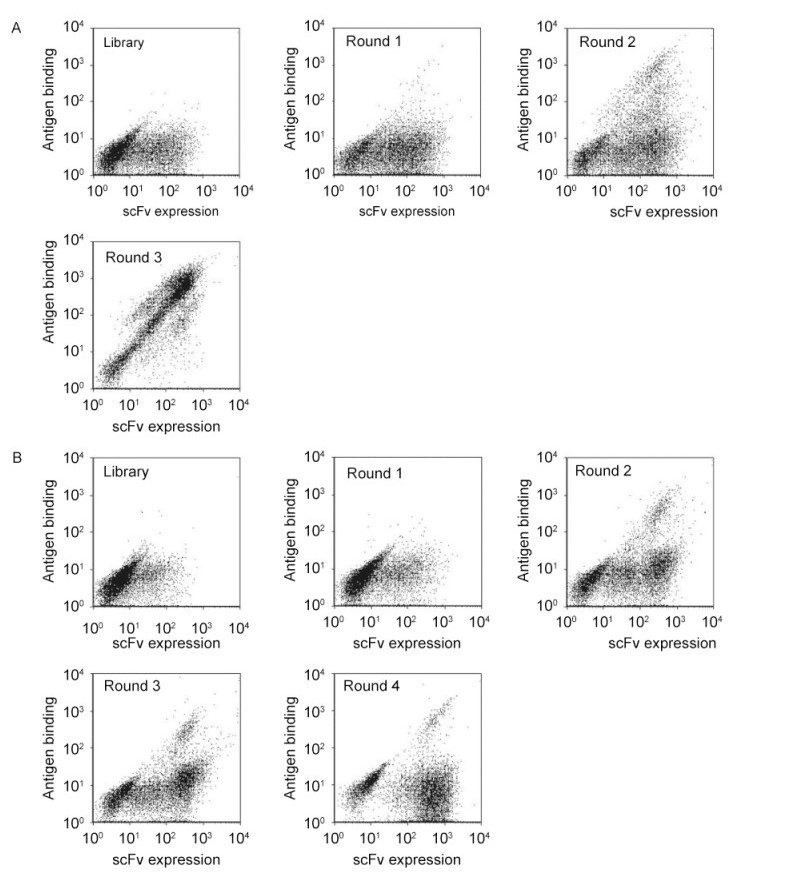
Figure 1. FACS analysis of scFv (A) and scFab (B) from yeast libraries for panning. Yeast cells were labeled with mouse anti-c-Myc antibody followed by goat anti-mouse dye, as well as biotinylated SP62 peptide followed by streptavidin-dye. Then, yeast cell were analyzed by flow cytometry. Yeast cells expressing scFv (A) and scFab (B) were selected for 3 and 4 rounds, respectively.
To further characterize the selected antibodies(3A2a and 3A5a), they were expressed as scFv fragments. ELISA showed that 3A2a and 3A5a specifically bound to MPER peptide SP62 with relatively high affinity(EC50 values of 2 nmol/L and 7 nmol/L)(Figure 2A, 2B), while no significant binding to scrambled SP62 peptide(SCR)was observed(Figure 2C, 2D).
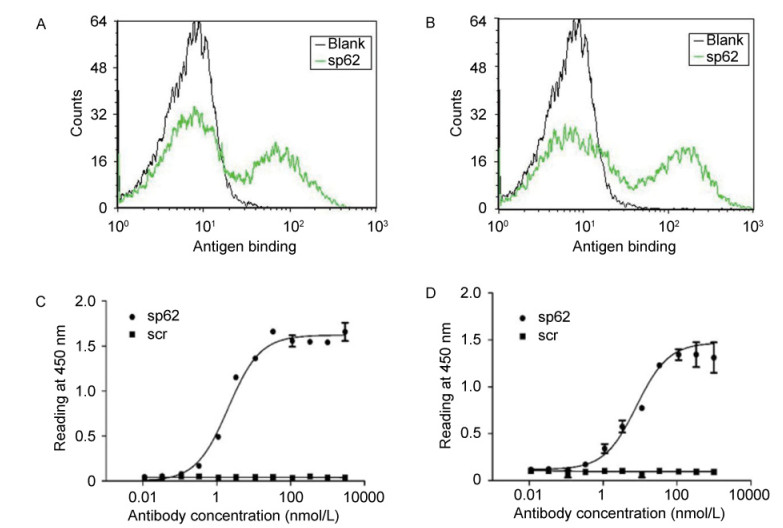
Figure 2. Specific binding of 3A2a and 3A5a scFvs to peptide SP62 (A, B). Yeast cells expressing 3A2a (A) and 3A5a (B) were treated with biotinylated SP62 peptide (green) followed by streptavidin-dye staining and analyzed by flow cytometry. The blank histogram indicates staining without antigen. Three-fold serially diluted 3A2a (C) and 3A5a (D) scFvs were added to 96-well plates coated with SP62 peptide (●) and control scrambled (SCR) peptide (■). Bound Abs were detected using HRP conjugated to anti-Flag antibody and 2, 2'-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) as substrate.
-
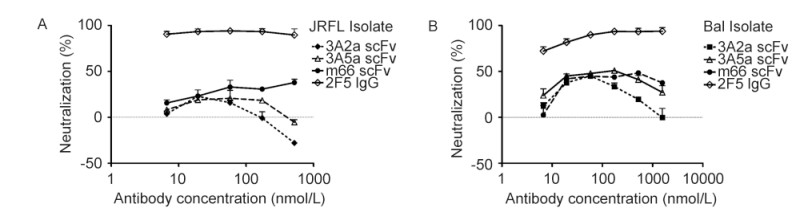
To investigate whether the antibodies were capable of neutralizing HIV-1 primary isolates, we used viruses pseudotyped with Envs from HIV-1 isolates JRFL and Bal, representing clade B. Neither of the antibodies exhibited strong neutralizing activity(Figure 3). Both 3A2a and 3A5a antibodies exhibited weaker neutralizing activity than m66 Fab. 3A2a and 3A5a could even enhance viral infection by JRFL when these antibodies were at high concentrations. The m66 antibody was previously identified as a bnAb 2F5-like antibody from an HIV-1 patient(Zhu et al., 2011). The positive control antibody 2F5 IgG showed efficient neutralization activity.
-
The epitopes of four well-known broadly neutralizing MPER-specific antibodies, 2F5, 4E10, Z13 and m66.6, include the MPER of gp41. They bind to linear peptide sequences located in the MPER. To find out whether our isolated antibodies bound to the same epitope as 2F5 antibody, we performed competition flow cytometric assays between 3A5a-displayed yeasts and 2F5 antibody or a control antibody(m102.4)against Nipah virus and Hendra virus(Zhu et al., 2008). The yeast cells competitively decreased the binding of 2F5 antibody to MPER peptide SP62, but not that of the irrelevant(negative control)antibody m102.4(Figure 4A). We also measured the binding of soluble scFv fragments of 3A5a in competition ELISA with 2F5 antibody. 3A5a significantly blocked the binding of 2F5 IgG to SP62 peptide(Figure 4B). These data implied that antibody 3A5a shared the same, or an overlapping, epitope with 2F5 antibody.
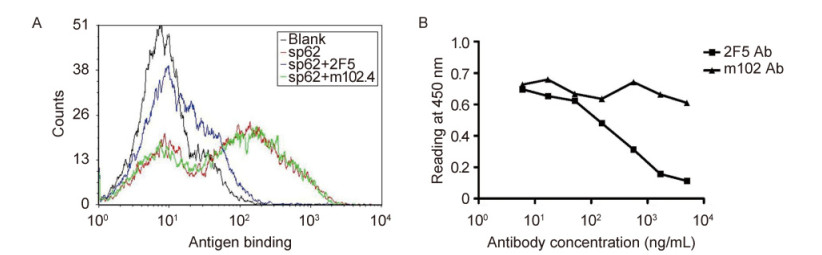
Figure 4. Competition assay of 3A5a scFv and 2F5 IgG for binding to SP62 peptide. (A) Enriched yeast cells from the scFv library were incubated with biotinylated SP62 peptide (red), a mixture of SP62 and 2F5 IgG (blue), or a mixture of SP62 and m102.4 IgG (green) and then stained with R-phycoerythrin conjugated streptavidin. Cells were detected by flow cytometry. (B) Mixtures of 3A5a scFv and 2F5 IgG (■) or control m102.4 IgG (▲) were added to 96-well plates coated with SP62 peptide. Bound Abs were detected using HRP conjugated to anti-Flag antibody and ABTS as substrate.
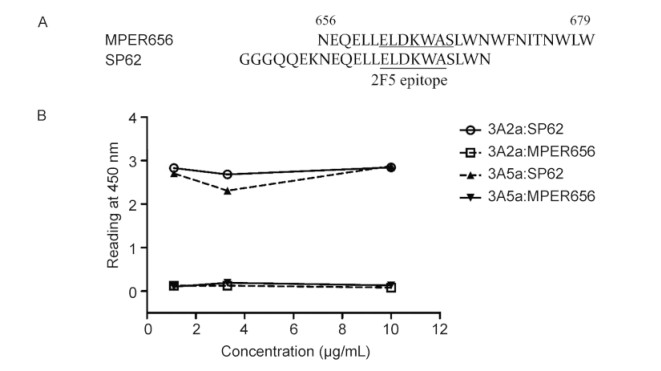
To further localize the antibody epitopes, we measured antibody binding to the MPER656 peptide that comprises the 2F5 and 4E10 epitopes but lacks several residues(aa652-655)that are present at the N-terminus of SP62(Figure 5A). Both 3A2a and 3A5a bond SP62 but not MPER656(Figure 5B). Thus, 3A2a and 3A5a lost binding activity on deletion of the sequence QQEK. Therefore, we suggest that the epitope of the weakly-neutralizing antibodies(mAbs)3A2a and 3A5a requires the 2F5 epitope(aa662-667) and, additionally, gp41652-655.
Generation of scFv and scFab yeast libraries
Selection of yeast surface displayed scFv and scFab libraries with MPER peptide
The selected antibodies weakly neutralized HIV-1 isolates
Characterization of the antibody epitopes
-
An effective HIV-1 vaccine should elicit broadly neutralizing antibodies targeting the HIV Envs, such as glycan shield, CD4 binding site, the coreceptor binding site, Env fusion intermediates, and MPER(Lin and Nara, 2007; Phogat and Wyatt, 2007). However, most of the HIV-1 vaccines based on unmodified Envs do not efficiently induce high-titer bnAbs to combat the highly variable viruses in human clinical trials.
In recent years, the very conserved gp41 MPER has gained attention as a new vaccine immunogen. However, the gp41 MPER epitopes are normally evanescent or poorly presented in vivo(Chen et al., 2010). In addition, the HIV-1 gp41 frequently induces non-neutralizing 2F5-like antibodies in HIV-1 patients. This raises the question of how the binding properties of gp41 non-neutralizing antibodies differ from those effective bnAbs. It is important to stabilize and present the 2F5 epitope in its proper structural configuration(Holl et al., 2014). The rational modification of MPER vaccines can prolong the exposure of the gp41-neutralizing epitopes, thus allowing for a longer time for MPER antibody binding.
In a previous report(Zhu et al., 2011), we described two 2F5-like antibodies(m66 and m66.6)that target the core 2F5 epitope(664DKW666) and two additional upstream residues(L660, L663). Neutralization of HIV-1 by m66.6 was more effective than that by m66, but weaker than that by 2F5. In this study, we described two 2F5-like antibodies(3A2a and 3A5a)targeting the highly conserved regions of gp41 which are in very close proximity to the 2F5 binding site. The binding profile of 3A2a and 3A5a to MPER peptides was very similar to that of m66. All of them showed high affinity for the SP62 peptide. Similar to m66, we observed that the localization of two 2F5-like antibodies required the 2F5 epitope(aa662-667) and several additional residues at the N-terminus of SP62. The difference was that the epitopes of 3A2a and 3A5a involve aa652-655 whereas the epitope of m66 involves aa660 and aa663. These results strongly indicate that the 3A2a and 3A5a epitope overlaps with that of 2F5, but the two epitopes are different.
m66 was less effective than 2F5 in a neutralization assay of HIV-1 primary isolates. Despite similarities to m66, the two 2F5-like antibodies identified here exhibited weaker HIV-1-neutralizing activity than m66. They gradually lost the ability to block the viral infection at high antibody concentration. We speculate that these antibodies might aggregate at high concentration due to low stability, resulting in non-specific binding to the cell surface. One could hypothesize that the possible high immunogenicity of the conserved non-neutralizing epitopes adjacent to the 2F5 epitope could further induce abundant non-neutralizing antibodies and shield neutralizing epitopes from immune system responses. In addition, these antibodies could directly prevent the detection of some neutralizing antibodies. Therefore, rational design of immunogens based on MPER would exclude the region aa652-657. These data also have important implications for reducing the immunogenicity of unwanted epitopes when the MPER region is used as an immunogen.
-
This work was supported by the Natural Science Foundation of Guangdong(No. 2015A030313741), the National Natural Science Foundation of China(No. 31440041), Shenzhen Peacock Innovation Plan Fund(No. KQCX20140520154115029), Shenzhen Knowledge Innovation Program(No. JCYJ20140901003939 026), and Novo Nordisk A/S -Chinese Academy of Sciences Research Fund(No. NNCAS-2013-9). We are grateful to Dr. Dimiter S. Dimitrov, National Cancer Insitute, NIH, for his expertise and advice.
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
DL and JL conceived and executed experiments. TX conceived and executed experiments related to antibody purification. JC conceived of and executed experiments related to library screening. LW conceived and co-wrote the manuscript. QZ supervised all aspects of the investigation and co-wrote the manuscript.














 DownLoad:
DownLoad: