HTML
-
Citrus tristeza virus (CTV), one of the most economically important citrus tree viruses, is distributed worldwide and has caused serious economic losses in the citrus industry (Sagheer et al., 2012). CTV belongs to the genus Closterovirus in the family Closteroviridae and has a long flexuous virion containing an approximately 20-kb positive-sense single-stranded RNA genome (Bar-joseph et al., 1989; Karasev et al., 1995). The virus has a host range restricted to most species of the family Rutaceae (Roistacher, 1991). CTV can cause disease symptoms including seedling yellowing, stem pitting, stunting, small fruit size, decline, and death (Sagheer et al., 2012; Moreno et al., 2008). CTV has been disseminated to citrus-growing zones through the exchange of in-fected buds and saplings. Subsequent spread by aphids and grafting during citrus propagation causes local epidem-ics (Bar-joseph et al., 1989; Moreno et al., 2008). The virus genome encodes two capsid proteins (CPs): the 25-kDa major CP encoded by the seventh open reading frame (ORF7) encapsidates approximately 95% of the helical nucleocapsid, whereas the 27-kDa minor CP encoded by ORF6 encapsulates the rest of the CTV genome and appears at one end of the capsid (Febres et al., 1997; Sekiya et al., 1991).
The most destructive outbreak of CTV first occurred in Argentina in 1930 and then moved to Brazil in 1937. From 1938 to 1980, epidemics were successively reported in Americas, Africa, and Europe. Outbreaks also occurred in Cyprus in 1989, Cuba in 1992, Mexico in 1995, the Dominican Republic in 1996, and Italy in 2002 (Bar-joseph et al., 1989; Davino et al., 2003; Garnsey et al., 2000; Gottwald et al., 2002; Moreno et al., 2008). CTV has been reported in various regions of China, and it has considerably reduced citrus production (Zhou et al., 1997). Therefore, a number of techniques have been established for detection and diagnosis of CTV, such as biological indexing (Cambra et al., 1990), serological assays (Bertolini et al., 2008; Cambra et al., 1990, 2000b; Permar et al., 1990), RT-PCR (Huang et al., 2004; Hung et al., 2000; Mehta et al., 1997), RT-nested PCR (Olmos et al., 1999), and real-time RT-PCR (Ananthakrishnan et al., 2010; Bertolini et al., 2008; Saponari et al., 2008). Current methods for CTV detection and diagnosis in China mainly include RT-PCR and biological indexing (Zhou et al., 2007).
Serological methods are the most convenient and cost-effective methods for large-scale screening (Wu et al., 2014). Tissue print enzyme-linked immunosorbent assay (ELISA) was developed to detect CTV without sample preparation (Cambra et al., 2000; Garnsey et al., 1993). Cambra et al.(1991)compared the sensitivities of different immunosorbent assays for CTV using CTV-specific monoclonal and polyclonal antibodies and found that triple antibody sandwich (TAS)-ELISA was the most sensitive, detecting as little as 0.1 ng/mL viral protein. Three CTV-specific monoclonal antibodies (MAbs) were obtained by Vela et al.(1986), and each MAb recognized all tested 23 strains of CTV with uniform reactions. Furthermore, Wu et al.(2014)identified three li-near epitopes (aa 48-63, 97-104, and 114-125) using bac-terially expressed truncated coat proteins and 10 MAbs against the native virions of CTV stem pitting isolate S4. Effective serological assays for detecting CTV would provide material and technical support for diagnoses, epidemiological analyses, and establishment of scientific prevention and control systems for CTV in China. However, reliable serological detection assays depend largely on antibody specificity and sensitivity.
In this study, four highly sensitive and specific MAbs for CTV were obtained through conventional hybridoma technology. Dot-ELISA, Tissue print-ELISA, and TAS-ELISA were developed for reliable and sensitive detection and diagnosis of CTV in China.
-
A stem pitting isolate of CTV from a Chongqing grove was identified with RT-PCR and biological indexing (Zhou et al., 2007) and maintained in a greenhouse in the authors' laboratory. Isolates of Citrus tatter leaf virus (CTLV) and Citrus yellow vein clearing virus (CYVCV) from Chongqing mandarin groves were identified by RT-PCR and nucleotide sequencing and then maintained in a greenhouse. The grove citrus samples including young stems and leaves of mandarin and orange trees were collected from Chongqing Municipality, Jiangxi Province, and Zhejiang Province in China during 2013-2014 and stored at -80 ℃.
-
Based on the nucleotide sequence of the major CP gene of the CT11 isolate of CTV in GenBank (accession no. EU 693526), the forward primer CP-F (5'-CTGCTACCATGGACGACGAAACAAAGAAATTG-3', with Nco Ⅰ restriction site underlined) and the reverse primer CP-R (5'-TTGTAGCTCGAGACGTGTGTTGAATTTCCCAAG-3', with Xho Ⅰ restriction site underlined) were designed for the gene cloning. Total RNA was extracted from CTV-infected citrus leaf samples using TRIzol reagent (Gibco-BRL, Gaithersburg, MD, USA), and the major CP of CTV was amplified by RT-PCR using the designed primers and the SuperScriptTM Ⅲ One-Step RT-PCR System (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The amplified major CP gene was purified by agarose gel electrophoresis, digested with Nco Ⅰ and Xho Ⅰ, and then subcloned into the His-tagged prokaryotic expression vector, pET-28a. The sequence and orientation of the ORF of the major CP gene in the recombinant expression vector were confirmed by nucleotide sequence analysis. The resulting recombinant expression vector pET-28a-CP was transformed into Escherichia coli BL21(DE3) cells. The major CP fusion protein was expressed, purified, and analyzed as described by Wu et al.(2009).
-
Using the expressed major CP of CTV as the antigen, PAbs were prepared as previously described (Wu et al., 2009). BALB/c mice were purchased from the Shanghai Laboratory Animal Center of the Chinese Academy of Sciences (certificate of animal quality: Zhong Ke Dong Guan No. 003). The animal experiments were performed in accordance with the Principles of the Helsinki accords and approved by the Animal Experimentation Ethics Committee of Zhejiang University, Hangzhou, China. The purified recombinant major CP protein was used as the immunogen to immunize five 8-week-old BALB/c mice as described previously (Wu et al., 2011). The hybridomas and MAbs were obtained as previously described (Wu et al., 2013). Titer determination, isotyping, specificity analysis, and purification of MAbs were carried out as described previously (Wu et al., 2011). The specificity and sensitivity of MAbs was determined by Western blot and TAS-ELISA (Wu et al., 2007).
-
TAS-ELISA for CTV detection was set up using the prepared rabbit PAb against the recombinant major CP of CTV, the prepared MAb, and alkaline phosphatase (AP)-conjugated goat anti-mouse IgG as the capture antibody, primary antibody, and enzyme-labeled secondary antibody, respectively. The TAS-ELISA standard protocol was performed as described by Wu et al.(2007). Citrus leaves were ground in liquid nitrogen and ground further in 0.01 mol/L phosphate-buffered saline (PBS), pH 7.4(100 mg leaf tissues in 2 mL PBS). Negative and posi-tive controls were the crude extracts from healthy and CTV-infected citrus leaves, respectively.
-
Nitrocellulose membranes were used as a sample support in dot-ELISA and Tissue print-ELISA for CTV detection. Phalanx tests were performed to determine the working dilutions of MAb and AP-conjugated goat anti-mouse IgG (Shang et al., 2011). Dot-ELISA protocols were carried out as described previously (Wu et al., 2014), and Tissue print-ELISA was performed as described by Shang et al.(2011). Tissue prints for Tissue print-ELISA were prepared by transversely cutting a rolled tender leaf or a young stem with a blade and gently pressing the freshly cut surface onto nitrocellulose membranes for 3 s. Negative and positive controls were the healthy and CTV-infected citrus tissues, respectively. Positive samples developed purple color within 10-20 min, whereas negative samples remained green.
-
Total RNA was extracted from citrus leaf samples using TRIzol reagent (Gibco-BRL), and the major CP gene of CTV was amplified by RT-PCR using the designed CP-F and CP-R primers and the SuperScriptTM Ⅲ One-Step RT-PCR System (Invitrogen) following the manufacturer's instructions.
Virus sources
Prokaryotic expression and protein purification
Preparation of polyclonal antibodies (PAbs) and MAbs
TAS-ELISA
Dot-ELISA and Tissue print-ELISA
RT-PCR
-
The 672-bp major CP gene of a CTV stem pitting isolate collected and identified from a citrus grove in Chongqing, China (Zhou et al., 2007) was amplified by RT-PCR and then inserted into the prokaryotic expression vector pET-28a. Sequencing analysis confirmed that the nucleotide sequence and the orientation of the ORF of the major CP in the recombinant expression vector pET-28a-CP were correct. E. coli BL21(DE3) cells harboring pET-28a-CP expressed an approximately 29-kDa recombinant fusion protein with IPTG induction, which coincided with the predicted molecular weight of the 6×His-tagged major CP (Figure 1A). Western blot analysis using an anti-His MAb (Roche, Indianapolis, IN, USA) as the primary antibody revealed a 29-kDa protein band in lanes with ly-sate from E. coli BL21(DE3) cells harboring the pET-28a-CP and purified recombinant protein (Figure 1B), indicating that the major CP recombinant protein was successfully expressed in E. coli BL21(DE3) and purified.
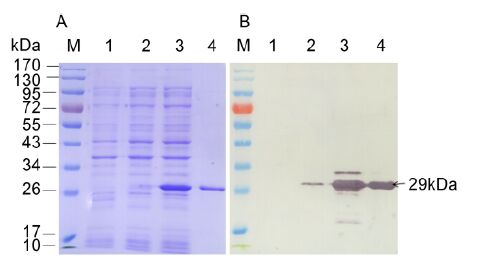
Figure 1. SDS-PAGE (A) and Western blot (B) anal-yses of the expressed major CP of CTV. Lane M: protein molecular weight marker. Lane 1: lysate of E. coli BL21 (DE3) harboring pET-28a with 0.5 mmol/L IPTG induction. Lanes 2 and 3: lysates of E. coli BL21 (DE3) harboring pET-28a-CP without or with 0.5 mmol/L IPTG induction, respectively. Lane 4: purified recombinant protein. An anti-His MAb was used to probe for the major CP recombinant protein in Western blot.
-
Four hybridoma lines (14B10, 14H11, 20D5, and 20G12) secreting MAbs specific for CTV were obtained after three cell fusion experiments and three cell cloning. Each hybridoma line was injected into pristane-primed BALB/c mice to produce ascitic fluid containing MAb. Isotypes and subclasses of all four MAbs belonged to IgG1, κ light chain. The titers of four MAbs determined by indirect ELISA ranged from 10-6-10-7, and IgG yields of MAbs in ascitic fluids ranged from 8.82 to 12.63 mg/mL (Table 1).

Table 1. Properties of CTV-specific monoclonal antibodies
The specificity of MAbs was first determined by Western blot. All four MAbs reacted strongly with the 25-kDa major CP of CTV in the crude extract from CTV-infected citrus leaf tissues but not with the crude extract from healthy citrus leaf tissues (Figure 2). An approximately 50-kDa protein band observed in CTV-infected tissues (Figure 2) was predicted to be a dimer of the major CP. The specificity of the MAbs with CTV-, CTLV-, and CYVCV-infected and healthy citrus leaf tissues was further tested by TAS-ELISA. All four MAbs reacted strongly with the crude extract from CTV-infect-ed citrus leaf tissues but not with the crude extracts from CTLV-, CYVCV-infected or healthy citrus leaf tissues (Figure 3), indicating that all four MAbs were specific for CTV.
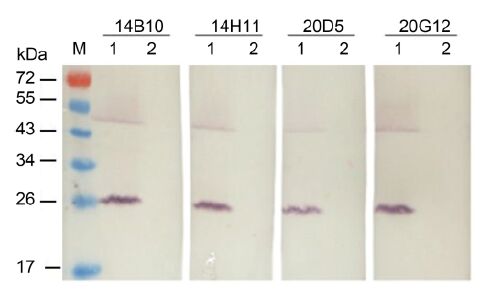
Figure 2. Specificity analyses of four MAbs against CTV by Western blot. M: protein molecular weight marker. Lanes 1 and 2: CTV-infected and healthy citrus leaf tissues, respectively. Each lane was loaded with 10 μg total protein.
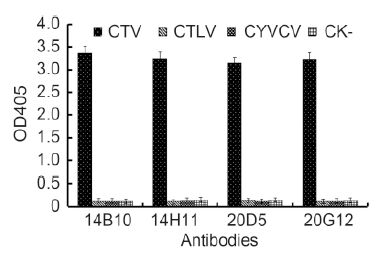
Figure 3. Specificity analyses of four MAbs by TAS-ELISA. Samples were diluted 1:20 (w/v, g/mL) and probed with MAbs diluted 1:5000 followed by AP-labeled goat anti-mouse IgG diluted 1:8000. Absor-bance value indicate the mean [±SD] from three independent assays at 30 min after adding the substrate at room temperature.
TAS-ELISA was also used to analyze the sensitivities of MAbs. The results of triplicate phalanx tests indicated that the PAbs (1:2000), the MAb (1:5000), and AP-conjugated goat anti-mouse IgG (1:8000) were optimal for TAS-ELISA. All four MAbs (14B10, 14H11, 20D5, and 20G12) could detect viruses in crude extracts of infected leaf tissue diluted up to 1:10, 240(w/v, g/mL), indicating that the MAbs and developed TAS-ELISA were very sensitive for CTV detection (Figure 4).
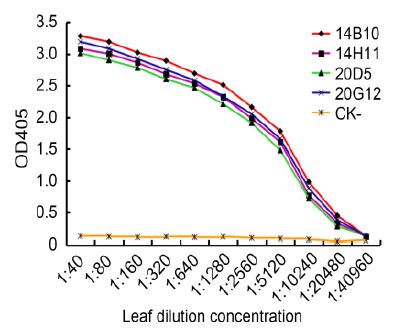
Figure 4. Sensitivity analyses of MAbs by TAS-ELISA. CTV-infected leaf crude extracts and healthy leaf crude extracts (CK-) were serially two-fold diluted in PBS from 1:40 to 1:40, 960 (w/v, g/mL) and probed with MAbs diluted 1:5000 followed by AP-labeled goat anti-mouse IgG diluted 1:8000. The OD405 absorbance values were measured 30 min after incubation with the substrate at RT. The OD405 values in figure indicate the mean of three independent assays.
-
Analytical phalanx tests demonstrated that 1:5000 and 1:8000 dilutions of MAb 14B10 and AP-conjugated goat anti-mouse IgG, respectively, were optimal in dot-ELISA for CTV detection. The developed dot-ELISA specifically detected CTV in infected citrus leaf tissues and was negative for CTLV-infected, CYVCV-infected, or healthy citrus leaf tissues (Figure 5A). Sensitivity anal-ysis revealed that the established dot-ELISA could detect viruses in infected citrus leaf extracts diluted up to 1:2560(w/v, g/mL) (Figure 5B).
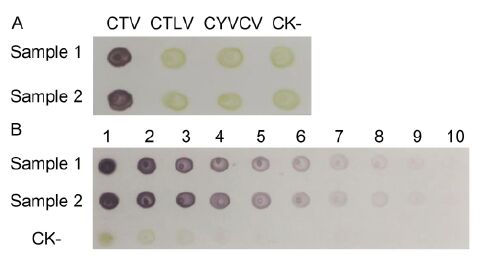
Figure 5. Specificity (A) and sensitivity (B) analyses of dot-ELISA. CTV-, CTLV-, and CYVCV-infected and healthy (CK-) citrus leaf tissue extracts were tested by dot-ELISA. 1-10: CTV-infected and healthy (CK-) citrus leaf tissue extracts were serially two-fold diluted from 1:10 to 1:5120 (w/v, g/mL), respectively. Purple indi-cates positive reactions, and green or no color indi-cates negative reactions. Samples 1 and 2 are two differ-ent CTV-infected citrus leaf tissues.
-
The working dilutions of MAb 14B10 and AP-conju-gated goat anti-mouse IgG in tissue print-ELISA were also determined by the phalanx test; the optimal dilutions of two antibodies in Tissue print-ELISA for CTV detection were 1:4000 and 1:8000, respectively. To identify a suitable tissue for CTV detection by Tissue print-ELISA, young fully expanded leaves and young stems of CTV-infected or healthy citrus trees were sectioned and printed on a nitrocellulose membrane. Purple spots were observed in prints of leaves and young stems from CTV-infected citrus trees, whereas green spots were observed in those from healthy citrus trees (Figure 6). Thus, both young leaves and stems of citrus trees were suitable for Tissue print-ELISA.
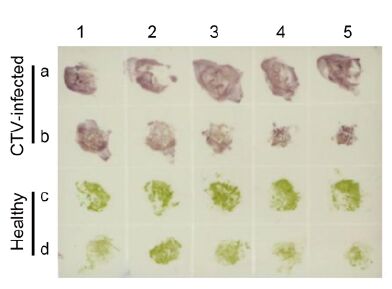
Figure 6. Detection of CTV in infected citrus tissues by Tissue print-ELISA. Young leaves and stems from CTV-infected citrus trees (a and b) or from healthy citrus trees (c and d) were tested. CTV in infected citrus tissues was probed with MAbs diluted 1:4000 followed by AP-labeled goat anti-mouse IgG diluted 1:8000. Purple and green colors indicate positive and negative reactions, respectively.
-
We screened 219 citrus samples collected from the Chongqing Municipality, Jiangxi Province, and Zhejiang Province of China during 2013-2014 for the presence of CTV using the developed serological assays. Sixty-eight of the 219 citrus samples were tested positive for CTV by Tissue print-ELISA, dot-ELISA, and TAS-ELISA (Table 2). The results from the three serological me-thods were consistent with those from RT-PCR except for one citrus sample collected in Zhejiang Province, which was tested negative in the serological assays but positive in RT-PCR (Table 2). DNA sequencing and alignment results confirmed that the PCR product was indeed amplified from the CTV genome. But, the coincidence rate of serological and RT-PCR test results reached more than 99.5%. Based on the survey results, we can infer that the MAbs we prepared can target a broad spectrum of CTV isolates in China.

Table 2. Detection of CTV in citrus groves
Expression and purification of the major CP of CTV
Production and characterization of MAbs against CTV
Dot-ELISA for CTV detection
Tissue print-ELISA for CTV detection
Survey of CTV in citrus groves
-
Detection and characterization of CTV are central to its prevention and control. Current CTV molecular detection methods mainly rely on PCR-based assays such as RT-PCR (Huang et al., 2004; Hung et al., 2000; Mehta et al., 1997), capture RT-nested PCR (Olmos et al., 1999), immunocapture RT-PCR (Cambra et al., 2002), and real-time RT-PCR (Ananthakrishnan et al., 2010; Bertolini et al., 2008; Saponari et al., 2008). Despite their high sensi-tivity, PCR-based methods are not suitable for routine large-scale CTV survey of citrus trees in groves (Wu et al., 2014). The most convenient and cost-effective me-thods for large-scale screening are serological assays, and many serological assays have been developed for CTV diagnosis and detection. MAbs specific for CTV have been successively produced (Nikolaeva et al., 1996; Ozturk et al., 2003; Permar et al., 1990; Vela et al., 1986), and double-antibody sandwich (DAS)-ELISA based on MAbs was developed for detecting CTV (Cambra et al., 1990; Garnsey et al., 1993; Permar et al., 1990). Using a uniform MAb, DAS-ELISA could detect all tested strains of CTV (Vela et al., 1986). Bertolini et al.(2008)reported that DAS-ELISA could detect CTV at dilutions up to 1:1000, whereas immunocapture-RT-nested PCR and real-time RT-PCR detected CTV targets at dilutions up to 1:5×106 and 1:109. Because the sensi-tivity of detection methods is especially important for a phloem-limited virus, the main drawback of serological assays is their lower sensitivity than molecular detection methods, which leads to false negative results. The sensi-tivity of serological assays is mainly determined by the qualities of the prepared antibodies.
In this study, using the recombinant major CP of CTV as the immunogen, we successfully prepared four highly sensitive and specific MAbs against CTV and used the MAbs and PAbs to develop three serological assays (dot-ELISA, Tissue print-ELISA, and TAS-ELISA) for detecting CTV in citrus groves. Our TAS-ELISA and dot-ELISA methods detected CTV in infected citrus leaf crude extracts at dilutions of 1:10, 240 and 1:2560, respectively. In particular, Tissue print-ELISA is a convenient, practical, and reliable method for detecting CTV in grove samples.
In our grove survey, the match rate between serological assays and RT-PCR was 99.5%. We sequenced PCR-amplified product of the citrus sample that showed posi-tive result in RT-PCR but negative result in the serological assays and confirmed that the PCR product was indeed amplified from the major CP ORF of CTV. Thus, the serological assays can give false negative reactions, but they demonstrated a high overall accuracy rate. Therefore, the serological detection approaches we developed herein have significant implications for large-scale surveys as well as long-term epidemiological or ecological studies of CTV.
-
This work was supported by Public Science and Technol-ogy Research Funds Projects of Agriculture (201203076-05).
-
All institutional and national guidelines for the care and use of laboratory animals were followed. All the authors declare that they have no conflict of interest.
-
ZL and ZC prepared the PAb and MAbs and carried out the immunoassays. XFW and CYZ performed the protein expression. ZL and JH participated in RT-PCR and the sequence alignment. XPZ and JXW conceived of the study, and participated in its design and helped to draft the manuscript.














 DownLoad:
DownLoad: