HTML
-
Baculoviruses are insect-specific viruses with a circular double-stranded DNA genome ranging in size from 80-180 kb (Lu et al., 2012). Two distinct types of virions have been identified during the infectious cycle of baculoviruses, namely budded virions (BVs) and occlusion-derived virions (ODVs). BVs mediate infection from cell to cell, while ODVs initiate oral infection in the insect midgut (Braunagel and Summers, 2007).
P33 is one of 37 core genes conserved in all sequenced baculovirus genomes (Rohrmann, 2014). P33 of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) was first identified as a flavin adenine dinu-cleotide (FAD)-linked sulfhydryl oxidase in enzymology studies (Long et al., 2009). The sulfhydryl oxidases such as the essential for respiration and viability (Erv) and quiescin-sulfhydryl oxidase (QSOX) family proteins are ubiquitous in eukaryotic cells and play important roles in protein oxidative folding, subcellular location, and respiratory chain assembly (Allen et al., 2005; Vitu et al., 2006; Alon et al., 2010). Sulfhydryl oxidase plays roles in virion assembly and propagation in various viruses (White et al., 2002). Previous studies showed that AcMNPV lacking the p33 gene could not produce infectious BVs and lacked multiple nucleocapsid ODVs (also referred to as MNPVs) instead only single nucleocapsid ODVs (SNPV) were observed in p33 deficiency virus.(Wu and Passarelli, 2010; Nie et al., 2011), suggesting that p33 is essential for virus infection and proliferation. P33 of both AcMNPV (AcP33) and Bombyx mori NPV (BmP33) displays a complex and unique quaternary structural arrangement that has not been previously observed in the sulfhydryl oxidase family (Hakim et al., 2011; Hou et al., 2012), suggesting that a novel catalytic mechanism of disulfide bond formation may exist in baculoviruses. The mechanisms underlying P33 viral infection and assembly remain unclear.
Here, we studied the function of P33 from Helico-verpa armigera nucleopolyhedrovirus (HearNPV), a SNPV that has been successfully used as a commercial pesticide against cotton worm (Sun and Peng, 2007).
HaP33 was expressed in Escherichia coli and purified by nickel affinity chromatography (Figure 1A). Sulfhydryl oxidase activity of HaP33 was measured as described by Long et al.(2009). A substrate of 1 mmol/L DTT was added to a 400-μL reaction mixture containing 4 μg purified HaP33 protein in PBS. The catalyzed reaction was performed at 37 ℃. At each time point, 20 μL reaction mixture was removed, mixed with 160 μL mea-surement buffer (2 mol/L urea, 100 mmol/L potassium phosphate, pH 7.5, 1 mmol/L EDTA), and 6 μL DTNB (Ellman's Reagent) to a final concentration of 100 mmol/L. After incubating the reaction for 30 s at room temperature, the optical density (A405 value) was determined by UV spectroscopy and the thiol content was calculated using an extinction coefficient of 13, 600 M-1cm-1. As expected, thiol content decreased as the reaction progressed compared to the control group (Figure 1B). The results showed that HaP33 was able to oxidize the thiol of DTT; therefore, the enzyme has sulfhydryl oxidase activity.
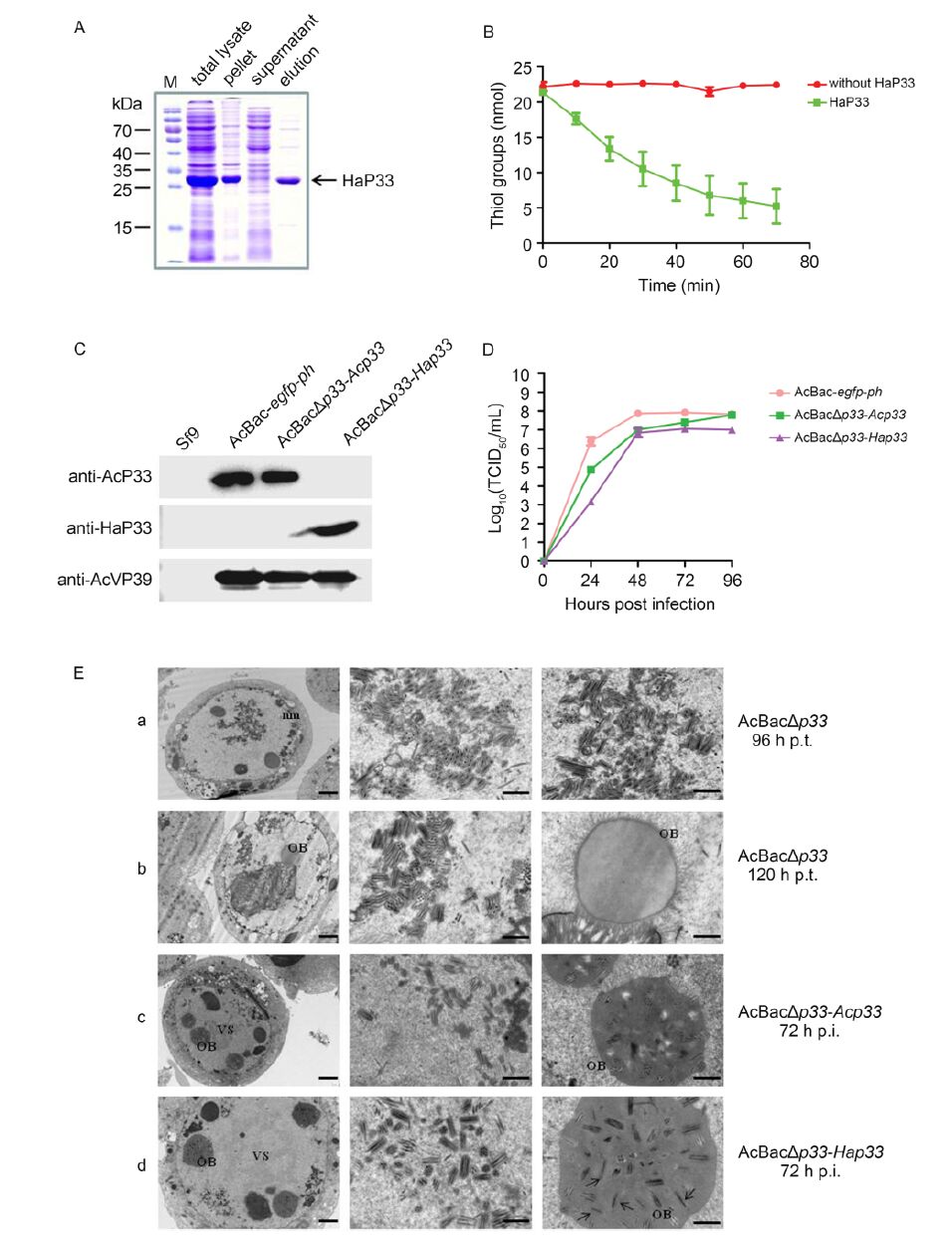
Figure 1. (A) Expression and purification of HaP33. HaP33 protein was expressed in the BL21(DE3) strain, purified by nickel affinity chromatography, and analyzed by SDS-PAGE. (B) Sulfhydryl oxidase activity assay. The reaction mixture containing purified proteins and the substrate DTT was incubated at 37 ℃; 20 μL aliquots were removed at the indi-cated times and the thiol groups were measured as described previously. Error bars indicate the standard deviation of three replicates. (C) Characterization of recombinant viruses by Western blotting. Cells were infected with the indicated viruses at an MOI of 5 and harvested at 48 h post-infection (p.i.). for immunoblotting with the indicated antibodies. (D) One-step grow curve assay of recombinant viruses. Sf9 cells were infected with vHaBac-egfp-ph, vAcBacΔp33-Acp33, and vAcBacΔp33-Hap33 at an MOI of 5 and the titers of supernatants collected at the indicated time points were determined by EPDA. Error bars indicate the standard deviation of three replicates. (E) Transmission electron microscopy analysis of Sf9 cells transfected or infected with recombinant viruses. Sf9 cells were transfected with vAcBacΔp33 (a and b) or infected with vAcBacΔp33-Acp33 (c) and vAcBacΔp33-Hap33 (d). Samples were harvested at 96 and 120 h post-transfection (p.t.) (a and b) or 72 h p.i. (c and d) for TEM analysis. The left panel shows the whole cell and the middle and right panels show the magnified sections of the whole cell. nm: nuclear membrane; OB: occlusion body; and VS: virogenic stroma. The arrows point to the single enveloped nucleocapsid embedded in the OB. The scale bars indi-cate 2 μm (left panel) and 0.5 μm (middle and right panel).
Deletion of the Acp33 gene suppresses the production of infectious BV and affects the formation of multiply enveloped ODV (Wu and Passarelli, 2010); thus, we substituted Hap33 for Acp33 to determine whether HaP33 is a functional homolog of AcP33. Acp33-knockout, Acp33-repaired, and Hap33-replaced bacmids were constructed and named bAcBacΔp33, bAcBacΔp33-Acp33, and bAcBacΔp33-Hap33, respectively. Among these bacmids, a large region (567 nt, 78762-79328 nt of AcMNPV) of the Acp33 open reading frame (total length of 780 nt) was deleted and replaced with a hsp70-egfp-SV40-Cm cassette, and egfp and polyhedrin (ph) were inserted at the ph locus to monitor virus infection. For the Acp33-repaired and Hap33-replaced bacmids, the Acp33 promoter was inserted upstream of Acp33 and Hap33 to initiate their transcription. The transfection and infection assays showed that infectious BVs could not be rescued from cells transfected with the Acp33 knockout bacmid, but could be obtained from those transfected with the Acp33-repaired and Hap33-replaced bacmids (data not shown); the rescued viruses were designated as vAcBacΔp33-Acp33 and vAcBacΔp33-Hap33, respec-tively. The expression of P33 in cells infected with recombinant viruses was detected by immunoblotting. Sf9 cells infected with vAcBac-egfp-ph, vAcBacΔp33-Acp33, and vAcBacΔp33-Hap33 at an multiplicity of infection of 5 were collected at 48 h post-infection (p.i.) and detected using antibodies against AcP33(anti-AcP33), HaP33(anti-HaP33), and AcMNPV VP39(anti-AcVP39). As shown in Figure 1C, no positive band was detected in uninfected cells, while a specific band with the predicted molecular weight was detected in infected cells, confirming that vAcBacΔp33-Acp33 and vAcBacΔp33-Hap33 were successfully constructed.
One-step growth curve analysis was conducted to determine the kinetic features of virus replication (Figure 1D). The replaced virus vAcBacΔp33-Hap33 exhibited similar viral growth curve kinetics to that of the control virus vAcBac-egfp-ph and repaired virus vAcBacΔp33-Acp33. However, the BV production of vAcBacΔp33-Hap33 was approximately 100-and 10-fold lower than that of vAcBacΔp33-Acp33 at 24 h p.i. and 48-96 h p.i, respectively. The repaired virus vAcBacΔp33-Acp33 also showed a 10-fold decrease in BV titers compared to that in the control virus vAcBac-egfp-ph at 24-48 h p.i., but reached similar levels at 72 and 96 h p.i. These re-sults indicate that HaP33 can partially rescue the function of AcP33 in generating infectious BVs.
Transmission electron microscopy analysis was performed to assess the impact of the replaced Hap33 on virion morphogenesis (Figure 1E). At 96 h post-transfection (p.t.), in most cells transfected with AcBacΔp33 bacmid, only single enveloped nucleocapsids were observed (Figure 1E-a). The results agreed with those of a previous study (Wu and Passarelli, 2010). However, at 120 h p.t., multiple enveloped nucleocapsids were observed, although they were not embedded into OBs (Figure 1E-b). Multiple enveloped nucleocapsids in AcP33-defective virus-transfected cells have not been previously reported (Wu and Passarelli, 2010). At 72 h p.i., cells infected with vAcBacΔp33-Acp33 exhibited the typical characteristics of infection, including multiply enveloped nucleocapsid assembly, ODV embedding, and mature OB formation (Figure 1E-c). In vAcBacΔp33-Hap33-infected cells, typical characteristics of infection were observed, with slightly more single-enveloped nu-cleocapsids in the OBs than that observed in vAcBacΔp33-Acp33-infected cells (Figure 1E-d). Thus, Hap33 can largely, but not perfectly, rescue the defects in morphogenesis caused by the absence of Acp33.
A previous study showed that AcP33 was a FAD-linked sulfhydryl oxidase that can oxidize sulfhydryl of DTT in vitro (Long et al., 2009). In this study, using a similar method, we showed that HaP33 displayed sulfhydryl oxidase activity in vitro (Figure 1B). The solution of purified HaP33 shows a yellow color (data not shown), suggesting that HaP33 contains FAD and is likely a FAD-linked sulfhydryl oxidase. In addition, we constructed a recombinant virus vAcBacΔp33-Hap33 in which Acp33 was replaced with Hap33, and the recombinant virus exhibited a similar viral growth pattern and viral particle morphology compared to the control and wild-type viruses. However, we also observed that the BV titer decreased by 10-fold, indicating HaP33 could only partially complement the function of AcP33. A similar result was reported when P33 from Trichoplusiani SNPV was used to replace AcP33(Clem et al., 2014). As HaP33 showed only 52% amino acid sequence identity with the AcP33, the two proteins may differ in their structure and function. Moreover, the heterologous expression of HaP33 in AcMNPV may cause some problems such as reduced protein yield or distinct intracellular localization. We determined the localization of HaP33 in vAcBacΔp33-Hap33-infected cells and found that its localization was similar to that of AcP33 in AcMNPV-infected cells (data not shown), indicating that subcellular distribution is not a major factor in the incomplete functional replacement.
Interesting, multiply enveloped nucleocapsids were observed in AcP33-deficient virus at 120 h p.t.(Figure 1E-b), which was not reported previously (Wu and Passarelli, 2010), suggesting that P33 is not the sole deter-mining factor of the SNPV and MNPV phenotypes. A recent review by Rohrmann (2014)suggested that many factors are involved in producing the S or M phenotype. Our results showed that in AcP33-deleted bacmid transfected cells, SNPVs were dominant during the early stage, but MNPVs were also detected in the late stage, suggesting that P33 plays an important role in ODV formation. As a sulfhydryl oxidase, P33 may affect the ODV enveloping process by regulating the disulfide bond formation of certain viral proteins. Our lab is currently studying proteins that are potential substrates of P33.
-
This work was supported by the grants from the National Science Foundation of China (No. 31570153, 31130058, 31321001 and 31400142) and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB11030400). The authors acknowledge the Core Facility and Technical Support of Wuhan Institute of Virology for technical assistance. The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: