-
Dear Editor,
Scylla serrata reovirus (SsRV), first discovered in China in 2004, is now known to be widespread in the mud crab (Scylla serrata) farming pond of China. (Chen et al., 2008). Since there are no effective treatments currently available for this viral pathogen, early diagnosis is the only way to prevent disease progression. Conventional methods for detection of SsRV are based on nested RT-PCR, one-step RT-PCR, and RT-loop-mediated isothermal amplification (RT-LAMP) (Guo et al., 2008; Chen et al., 2011). Even though PCR-based detection is sensitive, there are practical limitations that make such methods unsuitable for pond-side detection by farmers, not least the need for special instrument and professionally trained personnel. In the present study, we used polyclonal antibodies (pAb) raised against SsRV and monoclonal antibodies (mAb) specific for the SsRV capsid protein p29 to successfully develop a rapid and simple immunochromatographic strip for the facile detection of SsRV.
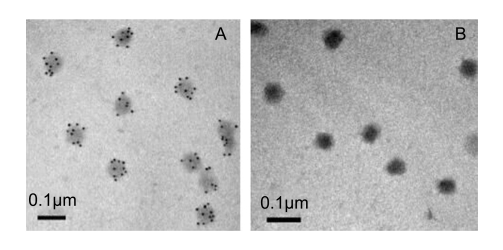
Antiserum against SsRV was generated by immunization of New Zealand White rabbits with purified SsRV (Chen et al., 2011), and was then purified by fast protein liquid chromatography. Preparation of mAb was performed as described previously (Zhou et al., 2005). Briefly, hybridomas of mouse myeloma cells SP2/0 and spleen cells derived from immunized BALB/c with purified recombinant his-tagged p29 (rHis-p29) (Yuan et al., 2016) were screened. After selection, one hybridoma clone stable secreting 5D4 specific to SsRV p29 was further purified by fast protein liquid chromatography. The ability of the mAb 5D4 to bind SsRV was determined by immunoelectron microscopy (IEM) as previously described (Leu et al., 2005). When a gold-labelled secondary antibody was used in conjunction with probe mAb 5D4, virions became coated with gold particles (Supplementary Figure S1A). Similarly, control experiments showed that gold particles were not observed on the outer surface of SsRV capsids when pre-immune mouse serum was used as the primary antibody (Supplementary Figure S1B).
Preparation of colloidal gold particles and colloidal gold-labelled antibody was performed as previously described (Zhang et al., 2006). mAb 5D4-conjugated colloidal gold particle were sprayed onto a glass fiber pad at a density of 3 μL/cm using a XYZ HM3030 Dispensing Platform to produce the conjugated pad. Rabbit anti-SsRV pAb (1 mg/mL) was then micro-sprayed onto a nitrocellulose membrane at a location that would turn into the test line (T) at a density of 1.5 μL/cm. Rabbit anti-mouse IgG antibody in PBS (1 mg/L) was then microsprayed on the same nitrocellulose membrane at a location that would turn into the control line (C) at a density of 1.5 μL/cm. After natural air drying at room temperature, the membrane was blocked with 2% (w/v) skim milk powder and dried at room temperature. The strips were assembled as described by previous report (Sithigorngul et al., 2007).
The specificity of the test strip for SsRV was evaluated using appropriate controls, such as white spot syndrome virus (WSSV), taura syndrome virus (TSV), hematopoietic necrosis virus (IHHNV) and uninfected mud crab. Individual hemolymph samples from uninfected S. serrata or SsRV-infected S. serrata collected in anticoagulant buffer (10% sodium citrate, pH 7) were mixed with MAS solution (27 mmol/L sodium citrate, 336 mmol/L NaCl, 115 mmol/L glucose, 9 mmol/L EDTA, pH 8.0) at a ratio 1:1 (v:v). Approximately 100 μL of each sample was added to the individual strip and incubated for 15 min. Red purple bands appeared within 15 min at both T and C lines when SsRV was detected, but only at the C line when it was not detected. Pleasingly, test strips were negative for controls, and only gave a positive result with SsRV-infected mud crab (Supplementary Figure S2A). This demonstrates that the strips can be used to detect SsRV and showed no cross-reactivity with WSSV, TSV and IHHNV.
To evaluate the sensitivity of the strips, hemolymph from S. serrata infected naturally with SsRV was serially diluted with MAS solution. Nucleic acids from 200 μL of each diluted sample were prepared using TRIzol reagent and tested for SsRV infection by one-step RT-PCR (Chen et al., 2011). These diluted samples were also used for SsRV strip tests as described previously. The lowest dilution of crab hemolymph from SsRV-infected mud crabs that gave a clear-cut positive test-strip result was 1:800 (Figure 1A). Comparison of strip test results with one-step RT-PCR showed a dim band for an amplicon of 180 bp at a dilution of 1×10–6 (Figure 1B). Thus, the strip test was 1000 times less sensitive than one-step RT-PCR for SsRV detection. Our test strips were comparable in sensitivity to previously reported yellow head virus (YHV) test strips that were also slightly inferior to PCR-based detection (Powell et al., 2006; Sithigorngul et al., 2007).
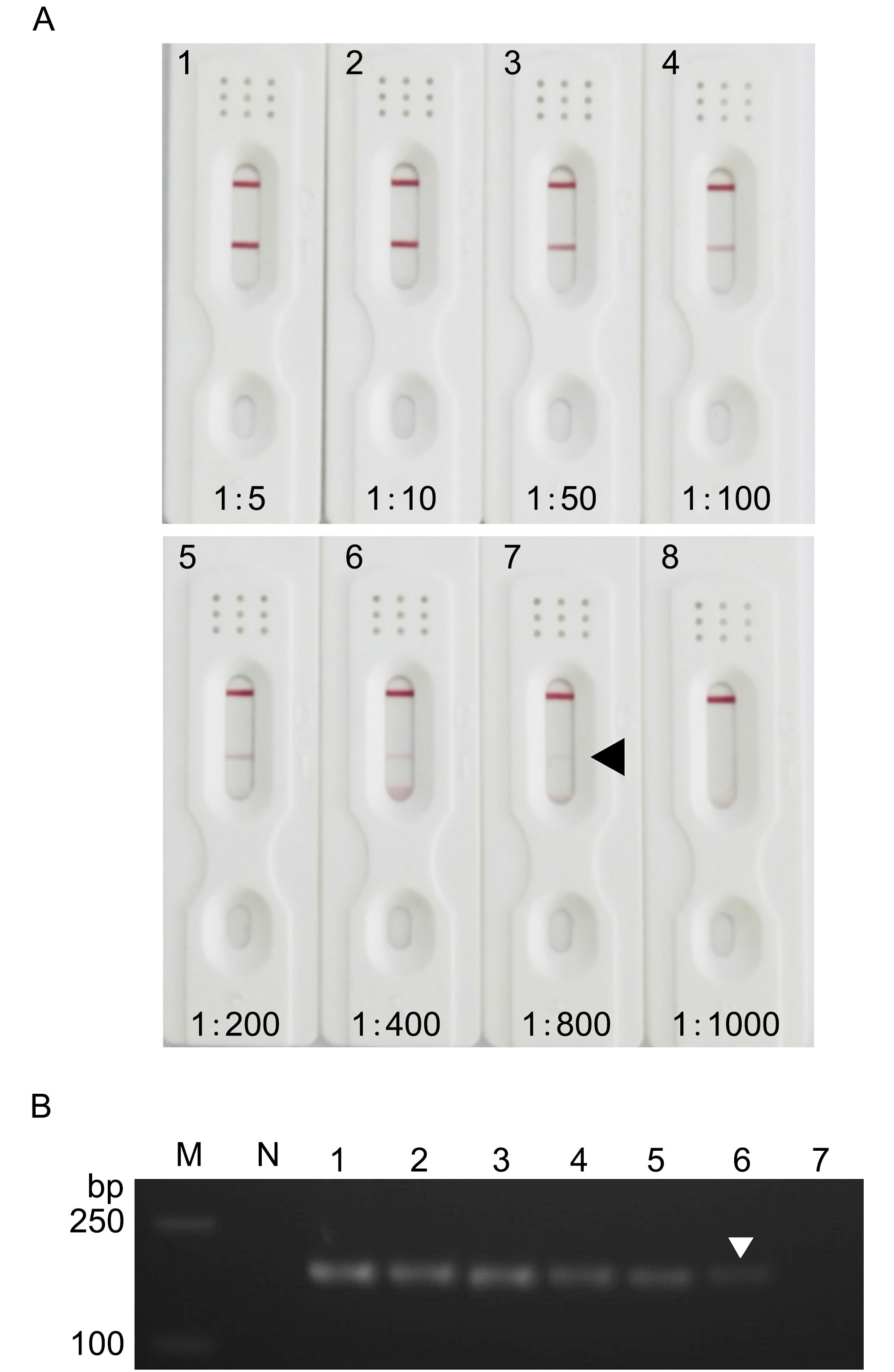
Figure 1. Comparison of detection sensitivity between immunochromatographic strips (A) and one-step RT-PCR (B). (A) Homogenates from SsRV-positive S. serrata following serial dilution with MAS solution and applied to strips at a volume of 100 μL per strip. N, SsRV-negative homogenate; lanes 1–8, serially diluted SsRV-positive homogenate at 1:5, 1:10, 1:50, 1:100, 1:200, 1:400, 1:800, and 1:1000, respectively. (B) RT-PCR of homogenates from SsRV-positive S. serrata following 10-fold serial dilution with MAS solution. Lane M, DNA markers; Lane N, SsRV-negative homogenate; lanes 1–7, serially diluted SsRV-positive homogenate from 1:10 to 1:10,000,000, respectively. Arrowheads indicate the lowest amount that could be reliably detected.
To evaluate stability, test strips that had been stored for 3, 6, 9, 12 and 15 months at 4 °C were tested repeatedly with SsRV-positive hemolymph and SsRV-negative hemolymph at 1:800 (three strips for each treatment) every 3 months and compared against a fresh batch of strips. Encouragingly, all test results were identical after 3 and 15 months of storage; positive and negative results remained as they were, and false positives were not detected. The results confirmed that strips could be safely stored for at least 15 months at 4 °C without loss of sensitivity.
To evaluate the reliability of the strip for on-site diagnosis, 35 S. serrata crabs from an SsRV-infected farm were used for specificity testing, of which 13 crabs showed classical gross signs of SsRV infection (erosive hepatopancreas), seven crabs demonstrated lightly atrophied hepatopancreas tissue, and the other 15 hepatopancreas samples were morphologically normal. All 20 crabs with erosive hepatopancreas or lightly atrophied hepatopancreas gave strong positive results with both the strip test and RT-PCR. For the rest of the crabs with normal hepatopancreas (15 samples), all gave negative strip test results, but eight samples gave positive results by one-step RT-PCR. Overall, the prevalence of positive samples was 80% (28 samples out of 35 samples) and 57% (20/35) for one-step RT-PCR and the strip, respectively (Table 1). Thus, compared with the RT-PCR-based reference standard, the overall sensitivity of the strip was 71% (20/28; Table 1). This result is similar to a previous report that compared YHV tests and RT- PCR in which the strip test failed to detect YHV in many samples lightly infected with YHV, but 100% of infected shrimps gave a positive result with PCR detection (Sithigorngul et al., 2007).
Number of samples (35) Strip One-step RT-PCR Number of positive samples 20 (57%) 28 (80%) Sensitivitya 71% Detection limit (dilution) 1:800 1:1,000,000 Detection time (min) 15 150 Price (USD) 3 12 Note: aCompared with the one-step RT-PCR-based reference standard. Table 1. Comparison of strip tests and one-step RT-PCR using S. serrata naturally infected with SsRV
The cost of various ingredients used in strip was compared with those used in one-step RT-PCR for a single reaction. Taking into account the cost of ingredients alone, one-step RT-PCR by conventional method costs USD 12, while the strip for one reaction costs only USD 3. Thus, the cost of strip is nearly 4 times less than one-step RT-PCR. In addition, one-step RT-PCR requires the use of expensive equipment and their operation requires professionally trained personnel. Strip, on the other hand, requires only a syringe for hemolymph sampling. Another advantage of the strip assay is that it gives results very quickly (within 15 min of sample preparation). By comparison, results from one-step RT-PCR require approximately 3 h excluding sample preparation (RNA extraction).
Although the lower detection limit of the strip is higher than that of one-step RT-PCR, the simplicity, cheapness and specificity of the strip would make it suitable for pond-side confirmation of SsRV infections in moribund crab at the early stage of disease outbreaks on mud crab farms, giving them time before severe crop loss for management decisions such as emergency harvest.
HTML
-
This research was supported by the Applied Research Programs of public welfare technology of Zhejiang Province of China (Grant No. 2014C32057), the innovative Research Team of Ningbo (Grant No. 2015C110018), and the Ningbo Municipal Natural Science Foundation (Grant No. 2016A610229). The authors declare that they have no conflict of interest. The study was approved by the Experimental Animal Ethics Committee of Zhejiang Wanli University, China. All the animal experiments were conducted in accordance with the guidelines of Zhejiang Wanli Univeristy for animal experiments.
Supplementary Figures are available on the websites of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.

















 DownLoad:
DownLoad: