HTML
-
Filamin belongs to the actin-binding protein family and other family members include talin, vinculin, and α-actin (Hartwig and Stossel 1975; Wang et al. 1975). Filamin is usually in the form of dimers that can link actin filaments into orthogonal networks or parallel bundles according to the actin:filamin ratio and the flexibility of the filamin isoform (van der Flier and Sonnenberg 2001). The N-terminus of filamin B encodes an actin-binding domain (ABD), which contains two calponin homology domains, known as CHD1 and CHD2. The ABD is followed by a series of 24 repeats that are folded into antiparallel β-sheets, with each repeat containing 96 amino acids (aa) (Gorlin et al. 1990). These repeats are interrupted by two hinge regions. Hinge Ⅰ is located between the 15th and 16th repeats (Xie et al. 1998; Xu et al. 1998), and hinge Ⅱ is located between the 23th and 24th repeats (Gorlin et al. 1990). It has been reported that filamin can interact with over 30 kinds of proteins, such as integrin (van der Flier et al. 2002), caveolin, smad, glycoprotein Ib-Ⅸ complex (Xu et al. 1998), insulin receptor and tissue factor (Stossel et al. 2001).
There are three types of filamin: filamin A, filamin B and filamin C. They are highly conserved in term of their nucleic acid sequences, but quite different in terms of their hinge area (Patrosso et al. 1994; Chakarova et al. 2000). Filamin A is found in all tissues, while filamin B (FLNB) mainly exists in the uterus, liver, thyroid and vascular endothelial cells. The gene encoding filamin B, which is also known as ABP278, is located in chromosome 3p14.3 (Brocker et al. 1999), and the protein has a variety of functions. For instance, it serves as a molecular scaffold for the type Ⅰ interferon-induced c-jun N-terminal kinase signaling pathway (Jeon et al. 2008), regulates chondrocyte proliferation and differentiation (Hu et al. 2014), and plays a key role in vascular endothelial growth factor-induced endothelial cell motility (del Valle-Perez et al. 2010). Filamin B mutations cause skeleton-related diseases including Larsen syndrome (LS) (Bicknell et al. 2007), atelosteogenesis Ⅰ phenotypes (AOI), atelosteogenesis Ⅲ phenotypes (AOⅢ) (Krakow et al. 2004), and Boomerang dysplasia (BD) (Bicknell et al. 2005).
Hepatitis B virus (HBV) infection can cause chronic hepatitis, which can develop into liver cirrhosis, liver fibrosis, and hepatocellular carcinoma (Cougot et al. 2005; Lupberger and Hildt 2007). HBV belongs to the Hepadnaviridae family (Beck and Nassal 2007), a family of small DNA viruses, and has four open reading frames (ORFs): S, P, X, and C. They encodes a variety of viral protein including viral envelope (pre-S1/pre-S2/S), core proteins (pre-C/C), viral polymerase, and HBx protein (Schadler and Hildt 2009).
Core protein is encoded by the C ORF and contains 183–185 aa (Scaglioni et al. 1997). Its N-terminal domain (NTD) consists of abundant α-helices (Dryden et al. 2006), and this domain regulates the assembly of core protein. The C-terminal domain (CTD) is rich in arginine and contains three SPRRR sequences (Salfeld et al. 1989). The CTD plays an important role in pre-genome/polymerase complex packing (Jung et al. 2014).
The HBV life cycle is a complex process. Core protein is involved in almost every step of the HBV life cycle. Firstly, HBV binds to Na+-taurocholate cotransporting polypeptide (NTCP) to enter the cytoplasm and releases the nucleocapsid (containing the viral genome) (Rabe et al. 2003; Yan et al. 2012). The function of core protein is to form the nucleocapsid, which moves to the nucleus and releases the viral genome (Kann et al. 2007). Later in the process, core protein packages the new made pre-genome/ polymerase complex (Bartenschlager et al. 1990) and this complex gradually forms the mature nucleocapsid (Lambert et al. 2007). Core protein promotes the transcriptional activity and stability of covalently closed circular DNA (ccc DNA), and interacts with the viral DNA to influence epigenetics (Guo et al. 2011). In the development of liver disease, core protein can induce tumor necrosis factor-α to mediate an immune response (Tzeng et al. 2014), sensitize hepatocytes to tumor necrosis factor-induced apoptosis (Jia et al. 2015), and regulates the replication of HBV (Liu et al. 2015).
Core protein plays an important role in the HBV life cycle, but there is little knowledge on how core protein interacts with host factors (Nguyen et al. 2008). Further study of the interaction between the virus and host is important to understand the HBV life cycle and discover new antiviral targets. A yeast two-hybrid system has shown that the CTD of core protein interacts with the CTD of human filamin B (Huang et al. 2000). This interaction has been further confirmed both in vitro and vivo (Huang et al. 2000). It is unknown where the interaction participates in HBV genome packaging, DNA replication, and virion morphogenesis. Filamin A can serve as an adaptor protein to connect the human immunodeficiency virus type Ⅰ (HIV-1) receptor and the cytoskeleton, promoting HIV infection (Jimenez-Baranda et al. 2007; Dotson et al. 2016). These studies suggest that filamin B may be involved in HBV infection, so the potential function between the interaction of core protein and filamin B needs to be studied.
In this study, we tested the interaction between filamin B and core protein in different cell lines using co-immunoprecipitation and co-localization immnofluoresence assay. And the influence of this interaction on HBV replication was examined at both RNA and protein levels using overexpression and knockdown strategies. After evaluation of the levels of HBV total RNAs, pre-genome RNA (pgRNA), and the expression levels of hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg), we found that the interaction of filamin B and core protein promotes HBV replication, impairing the interaction downregulates HBV transcription, replication and protein expression. Lastly, we identified the potential interaction sites in core protein. Taken together, these results strongly indicated that the interaction between filamin B and core protein plays an important role in regulating HBV replication. These data provide new insight into the relationship between HBV and host factors, which may provide new strategies for the treatment of HBV infection.
-
The pHBV1.3 (serotype adw, genotype B) plasmid was generated from the HBV genome (GenBank accession number JN406371) inserted into the pUC18 plasmid. The Core-HA plasmid contained the HBV core protein gene, was digested with Kpn Ⅰ/Hind Ⅲ and inserted into pXJ40-HA. Filamin B-GFP (designated FLNB-GFP) was provided as a generous gift from Professor Arnoud Sonnenberg (Netherlands Cancer Institute). We used the wild-type Core-HA plasmid (designated WT-HA) as a template to generate mutated versions of the Core-HA plasmid (designated M1-HA, M2-HA, M3-HA, M4-HA; each involving one of the four blocks of arginine residues), and the primers for the mutants are listed in Supplementary Table S1. Small interfering RNAs (siRNAs; siFLNB-1, siFLNB-2, siFLNB-3) and the negative control (siNC) were purchased from RiboBio (Guangzhou, China).

Table S1. Primer sequences used in the construction of mutants
-
HEK 293T cells, Huh7 cells and HepG2 cells (from the American Type Culture Collection [ATCC]) were maintained in Dulbecco's modified Eagle's medium (Gibco, Shanghai, China) supplemented with 10% fetal bovine serum (Gibco) at 37 ℃ and 5% CO2. Cells were seeded in 6-well plates (4 × 105 cells/well) or 12-well plates (2 × 105 cells/well) and grown to ~ 70% confluence at the time of transfection. The siRNA and plasmids were transfected into cells using Lipofectamine 2000 reagent (Invitrogen, Shanghai, China).
-
Cells grown in 10-cm2 culture dishes, and FLNB-GFP or GFP (10 μg) were co-transfected with the Core-HA plasmid (10 μg) using Lipofectamine 2000 reagent according to the manufacturer's protocol. Transfected cells were harvested at 48 h post transfection, then washed with PBS and lysed with 1 mL Nonide P-40 supplemented the 10 μL 1009 Cocktail (Biyuntian, Shanghai, China) and 10 μL 1009 PMSF (100 mmol/L) (Biyuntian). Cell lysates were centrifuged (13, 000 ×g) at 4 ℃ for 10 min to remove the nuclei, and then incubated with 40 μL proteinA/G (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and 8 μg antiGFP antibody at 4 ℃ overnight. After centrifugation, the sediment was washed three times with washing buffer (50 mmol/L Tris [pH 7.4] supplemented with 150 mmol/L NaCl).
-
Cells were lysed in radioimmunology precipitation assay (RIPA) buffer with SDS. The protein samples were then separated by SDS polyacrylamide gel electrophoresis (PAGE) and transferred onto a nitrocellulose membrane (GE Healthcare, Waukesha, WI, USA). The membrane was then blocked with skim milk and probed with antibodies. The antibodies were anti-HA (Santa Cruz Biotechnology), anti-GFP (Santa Cruz Biotechnology), anti-filamin B (GeneTex, CA, USA), anti-GAPDH (Sigma, St. Louis, MO, USA) and anti-tubulin (Sigma). The immunoblot signals were detected using an enhanced chemiluminescence (ECL) substrate (Bio-Rad, Shanghai, China).
-
Cells were cultured on glass coverslips and transfected with FLNB-GFP and Core-HA plasmids respectively, or co-transfected pair-wise, using Lipofectamine 2000 for 24 h. Cells were rinsed three times with PBS, fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.2% Triton X-100 for 15 min, and blocked with 5% bovine serum albumin in PBS for 30 min at room temperature. Then cells were then incubated with polyclonal rabbit anti-HA antibody (Santa Cruz Biotechnology), followed by 5 μg/mL Alexa 568 anti-rabbit antibody (Invitrogen) for 1 h at room temperature. At last, cells were permeabilized and stained with DAPI. Coverslips were mounted using Gel Mount (Biomedia), and the fluorescence was viewed with a Leica confocal microscope (LEICA Spectral Confocal TCS-SL, Serveis Cientı ´fico Te'cnics de la Universitat de Barcelona). Colocalization between filamin B and core protein was quantified by using Leica confocal microscope software. R values are shown as Pearson correlation coefficient ± SD averaged from 5 to 8 cells.
-
Quantitative real-time PCR analysis was performed to determine the level of HBV total RNAs and HBV pgRNA. Cells were grown in 6-well plates and transfected with the following plasmids: pHBV1.3, GFP, and pXJ40-HA; pHBV1.3, Core-HA, and GFP; pHBV1.3, FLNB-GFP, and pXJ40-HA; pHBV1.3, FLNB-GFP, and Core-HA. Total RNAs were isolated from cells using TRIzol Reagent (Invitrogen). The RNA (1 μg) was reverse transcribed with RT primer mix (oligo dT primer and random 6 mers) (TaKaRa). Quantitative PCRs were performed with SYBR Select Master Mix (Life Technologies) using the StepOne Real-Time PCR System (Applied Biosystems). The mRNAs levels were normalized to the GAPDH expression level.
PCR was performed at 95 ℃ for 3 min followed by 40 cycles at 95 ℃ for 15 s, 58 ℃ for 30 s, and 72 ℃ for 30 s. Each set of PCRs was performed in triplicate, and the CT values were obtained for each PCR. The ΔΔCT method was used to calculate the ratios of gene expression relative to the control set (pHBV1.3, GFP, and pXJ40-HA group) in the experiments. The primers used are list in Supplementary Table S2.

Table S2. Primer sequences used in quantitative real-time PCR
-
Cells were grown in 6-well plates and transfected with the following plasmids: pHBV1.3, GFP, and pXJ40-HA; pHBV1.3, Core-HA, and GFP; pHBV1.3, FLNB-GFP, and pXJ40-HA; pHBV1.3, FLNB-GFP, and Core-HA. The culture medium was collected at 48 h post transfection. The level of HBsAg and HBeAg in the culture medium was determined using ELISA kits (Beijing Wantai Biotech, Beijing, China) according to the manufacturer's instructions.
-
Cells were grown in 12-well plates and transfected with the following plasmids: pHBV1.3, GFP, and pXJ40-HA; pHBV1.3, Core-HA, and GFP; pHBV1.3, FLNB-GFP, and pXJ40-HA; pHBV1.3, FLNB-GFP, and Core-HA. For the Southern blot analysis, HBV DNA from intracellular core particles was extracted at 96 h post-transfection. Cells were lysed in 400 μL lysis buffer (50 mmol/L Tris–Cl [pH 7.4], 1% Nonidet P-40, 1 mmol/L EDTA) at 4 ℃ for 30 min and subsequently centrifuged (13, 000 ×g) at room temperature. The supernatant was then transferred to fresh tubes and incubated with 10 μL DNase Ⅰ (1 U/μL; Sigma) and 5 μL RNase A (10 mg/mL; Sigma) at 37 ℃ for 2 h. Subsequently, 20 μL EDTA (0.5 mol/L) was added to inactivate DNase Ⅰ, and the mixture was then incubated with 10 μL protease K (20 mg/mL Sigma) and 20 μL 20% SDS at 55 ℃ overnight. The digestion mixture was extracted with phenol/chloroform, and DNA was precipitated with ethanol and dissolved in 15 μL TE buffer (10 mmol/L Tris–HCl [pH 8.0], 1 mmol/L EDTA). All the extracted DNA was separated on 0.8% agarose gels. After standard denaturation and neutralization procedures, the DNA was transferred onto a positively charged nylon membrane (GE Healthcare, Waukesha, WI, USA) and probed with a digoxigenin (DIG)-labeled HBV RNA probe. The probe preparation and subsequent DIG detection were conducted using the DIG Northern Starter Kit (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer's instructions. pHBV1.3 plasmid was used as HBV DNA marker. HBV DNA marker bands are shown for relaxed circular DNA (rcDNA), double stranded DNA (dsDNA), and single stranded DNA (ssDNA). Ratios were quantified by gray analysis using GeneTools from Syngene software (GeneGnome5).
Plasmid Constructions and Filamin B siRNAs
Cell Culture and Transfection
Co-Immunoprecipitation Assay
Western Blot Analysis
Immunofluorescence
Quantitative Real-Time PCR Analysis
Enzyme-Linked Immunosorbent Assay (ELISA)
Southern Blot Analysis
-
It has previously been shown that filamin B interacts with HBV core protein. This interaction between the C-terminal region of filamin B and core protein has been demonstrated both by a pull-down assay in vitro and by a co-immunoprecipitation in vivo (Huang et al. 2000). To verify the interaction between filamin B and core protein, we used coimmunoprecipitation analysis in several cell lines. In this co-immunoprecipitation experiment, filamin B was tagged with GFP (designated FLNB-GFP), and core protein was tagged with HA (designated Core-HA). FLNB-GFP or GFP vectors were co transfected with the Core-HA plasmid into HEK 293T cells (Fig. 1). We used anti-GFP antibody to pull down FLNB-GFP and GFP. The aimed bands were detected by anti-GFP and anti-HA antibodies. The result demonstrated that FLNB-GFP could pull down Core-HA. In contrast, there was no interaction between GFP and Core-HA (Fig. 1). Similarly, this interaction was also detected in Huh7 cells and HepG2 cells (Fig. 1), which are both hepatoma cell lines. These data suggest that fulllength filamin B interacted with core protein in all three cell lines.
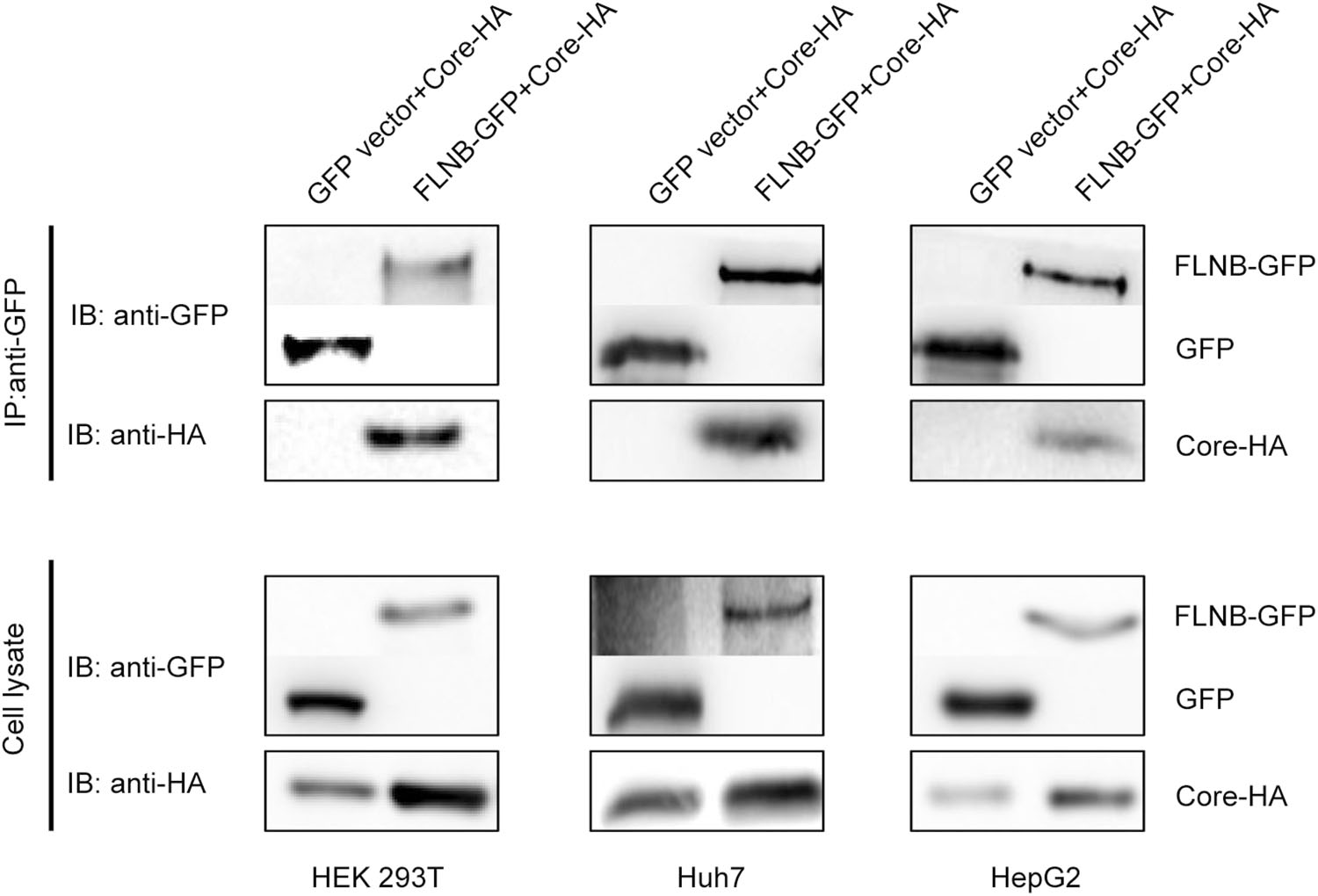
Figure 1. Filamin B interacts with core protein in different cell lines. Co-immunoprecipitation of filamin B and core protein is shown. Either FLNB-GFP plasmid or GFP vector (10 μg) was co-transfected with 10 μg Core-HA plasmid into HEK 293T, Huh7, and HepG2 cells. Filamin B was immunoprecipitated using GFP. Co-immunoprecipitated core protein was detected by immunoblotting with anti-HA antibodies (top). Western blot analysis of the precipitated filamin B and GFP using antiGFP antibodies is shown (top). Cell lysates before immunoprecipitation were probed with anti-GFP and antiHA antibodies (bottom).
-
To determine whether filamin B colocalizes with core protein in human cells, we performed confocal immunofluorescence microscopy. Filamin B is a cytoskeletal protein and wildly distributed across the whole cell, but it is concentrated more in the cytoplasm than the nucleus (Feng and Walsh 2004). Indeed, in cells without core protein expression, filamin B was mainly detected in the cytoplasm (Fig. 2A). The localization of core protein is related to the cell cycle (Yeh et al. 1993; Weigand et al. 2010) and the level of HBV replication (Chu et al. 1997; Serinoz et al. 2003), and it existed both in the cytoplasm and nucleus (Fig. 2B). We examined the colocalization of filamin B and core protein. In our study, filamin B and core protein expression plasmids were co-transfected pair-wise into HEK 293T cells, and a significant portion of core protein colocalized with filamin B in the cytoplasm (Fig. 2C). We also found colocalization in Huh7 and HepG2 cells (Fig. 2C). The colocalization rates were examined and Pearson correlation coefficient R values were 0.71, 0.83, and 0.79 for 293T, Huh7 and HepG2 cells, respectively. This revealed that filamin B colocalized with core protein in human cells, and further confirmed that filamin B interacted with core protein.
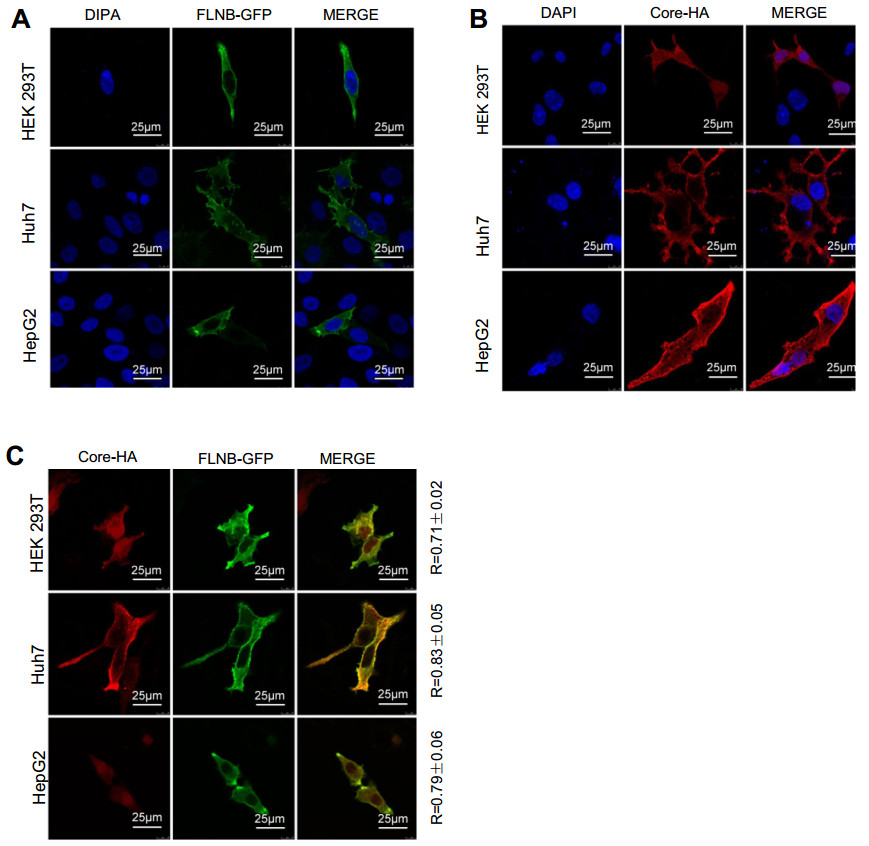
Figure 2. Filamin B colocalizes with core protein. (A) Confocal analysis of filamin B subcellular localization is shown in green. Cells were transfected with 2 μg FLNB-GFP plasmid. At 24 h post transfection, fixed cells were permeabilized and stained with DAPI. (B) Confocal analysis of core protein subcellular localization is shown in red. Cells were transfected with 2 μg of Core-HA plasmid. At 24 h post transfection, fixed cells were permeabilized and immunostained for core protein using anti-HA antibodies and fluorescent secondary antibodies. (C) Immunofluorescence analysis of the colocalization of filamin B and core protein is shown. Cells were transfected with 1 μg FLNB-GFP plasmid and 1 μg Core-HA plasmid. At 24 h posttransfection, fixed cells were permeabilized and immunostained for core protein using anti-HA antibodies and fluorescent secondary antibodies. Filamin B is shown in green (left), core protein in red (center), and colocalization in yellow (right). Scale bars represent 10 lm. Colocalization between filamin B and core protein was quantified using Leica confocal microscope software. R values are the Pearson correlation coefficient ± SD averaged from 5 to 8 cells.
-
Real-time PCR revealed that total RNAs and pgRNA of HBV were remarkably up-regulated 6–8-fold (pHBV1.3, FLNB-GFP, and Core-HA group) in comparison to the control (pHBV1.3, GFP, and pXJ40-HA group) in Huh7 and HepG2 cells (Fig. 3A, B). ELISA assay showed that the secretion level of HBeAg and HBsAg in the culture medium of Huh7 and HepG2 cells increased (Fig. 3C, D). Furthermore, Southern blot analysis suggested that the interaction significantly promoted HBV DNA replication (Fig. 3E). The control group (pHBV1.3, GFP, and pXJ40-HA) was set at a value of 1, and the other two groups (pHBV1.3, FLNB-GFP, pXJ40-HA group and pHBV1.3, FLNB-GFP, Core-HA group) increased 1.7–2.8-fold in comparison to the control group. The Western blot analysis indicated that filamin B was overexpressed. The real-time PCR and ELISA data showed that co-transfection with pHBV1.3 and Core-HA had no significant effect on HBV replication, so the Southern blot analysis did not involve this control experiment. These results indicated that the interaction between filamin B and core protein contributed to viral replication.
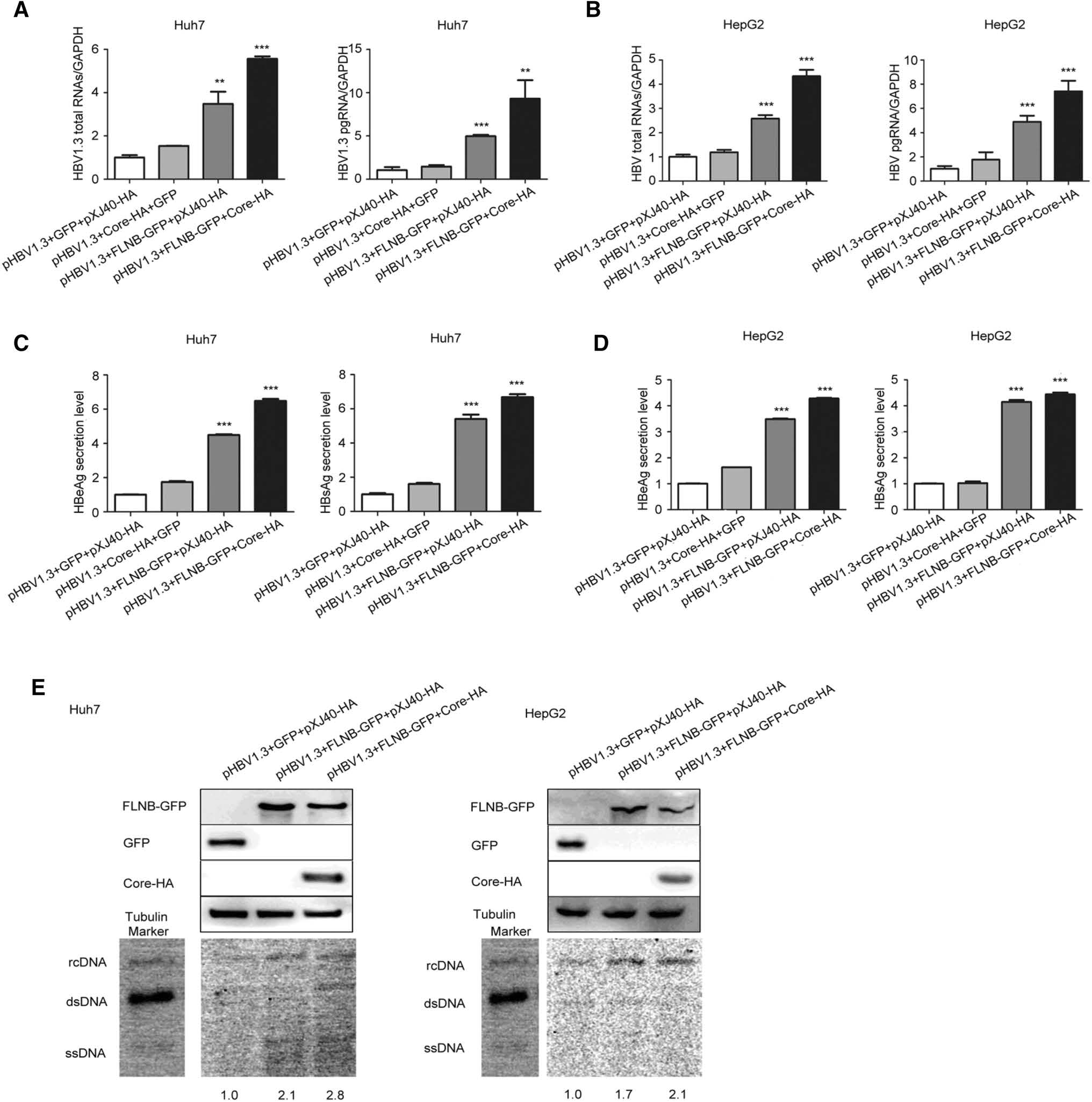
Figure 3. Interaction of filamin B and core protein promotes HBV replication. (A) Levels of HBV total RNAs and pgRNA in Huh7 cells were determined by real-time PCR. (B) HBV total RNAs and pgRNA levels in HepG2 cells were determined by real-time PCR. (C) Levels of HBeAg and HBsAg in culture medium supernatant of Huh7 cells were detected with ELISA. (D) Levels of HBeAg and HBsAg in culture medium supernatant of HepG2 cells were detected with ELISA. (E) HBV DNA was extracted at 96 h post-transfection and subjected to Southern blotting. Western blot showed that all proteins were overexpressed. Co-transfection with pHBV1.3, GFP and pXJ40-HA was used as a loading control. Ratios were quantified by gray analysis with GeneTools from Syngene software (GeneGnome5). HBV DNA marker bands are shown for relaxed circular DNA (rcDNA), double stranded DNA (dsDNA), and single stranded DNA (ssDNA). All results are shown as mean ± SD. (*P < 0.05, **P < 0.01, ***P < 0.001).
-
To further elucidate the role of the interaction in HBV replication, the Huh7 and HepG2 cells were transfected with filamin B-targeting siRNAs (siFLNB-1, siFLNB-2, and siFLNB-3) or siRNA negative control (siNC). The knockdown efficiency was detected using Western blotting, which indicated that all the siRNAs showed strong knockdown effects but siFLNB-2 was more effective than siFLNB-1 and siFLNB-3. This was consistent between Hun7 and HepG2 cells (Fig. 4A). We chose to use siFLNB-2 in our subsequent experiments. Real-time PCR results revealed that knockdown of filamin B remarkably reduced the level of HBV total RNAs and pgRNA in Huh7 and HepG2 cells (Fig. 4B, C). Consistently, the secretion level of HBeAg and HBsAg in culture supernatant (pHBV1.3 and siFLNB-2 group) decreased compared with the control cells (pHBV1.3 and siNC group) (Fig. 4D, E). In addition, the Southern blot results suggested that HBV replication decreased in filamin B knockdown cells (Fig. 4F). The gray analysis was executed by Syngene software (GeneGnome5), and the pHBV1.3 and siNC group was used as the control group and set at a value of 1. The other group (pHBV1.3 and siFLNB-2 group) decreased by 0.38–0.55-fold in comparison to the control group. The Western blot results indicated that filamin B was knocked down. Taken together, these data suggested that filamin B knockdown inhibits HBV replication.
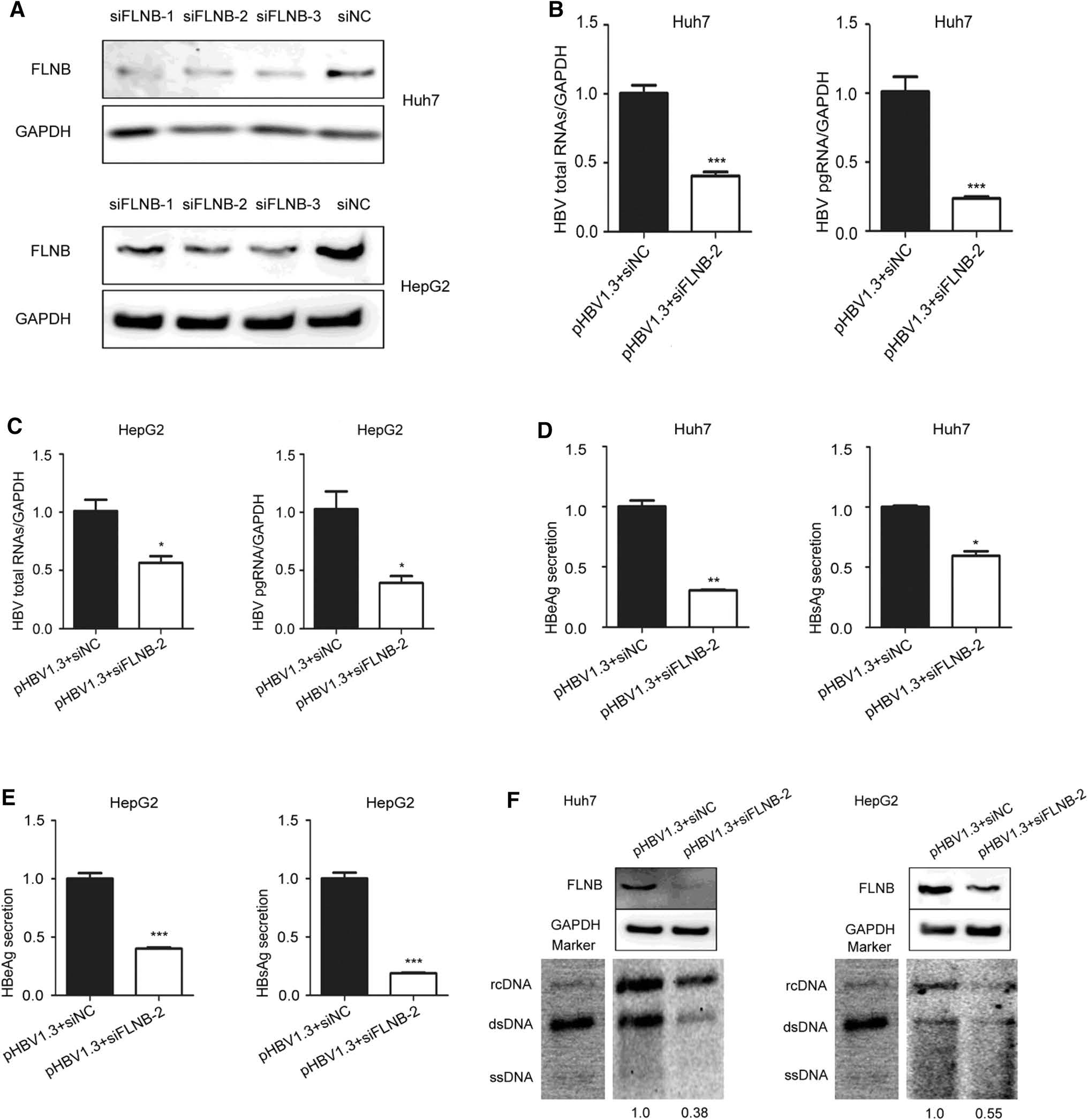
Figure 4. Filamin B knockdown inhibits the replication of HBV. (A) Huh7 and HepG2 cells were transfected with siNC, filamin B-targeting siFLNB-1, siFLNB-2 or siFLNB-3 separately. Western blot analysis of filamin B was used to confirm the silencing efficiency. (B) Levels of HBV total RNAs and pgRNA in Huh7 cells were determined by real-time PCR. (C) Levels of HBV total RNAs and pgRNA in HepG2 cells were determined by real-time PCR. (D) Levels of HBeAg and HBsAg in culture medium supernatant of Huh7 cells were detected with ELISA. (E) Levels of HBeAg and HBsAg in culture medium supernatant of HepG2 cells were detected with ELISA. (F) HBV DNA was extracted at 96 h post transfection, and then subjected to Southern blotting. Western blot showed that filamin B was successfully knocked down. Co-transfection with pHBV1.3 and siNC was used as a loading control. Ratios were quantified by gray analysis with GeneTools from Syngene software (GeneGnome5). rcDNA, relaxed circular DNA; dsDNA, double stranded DNA; ssDNA, single stranded DNA. All results are shown as mean ± SD. (*P < 0.05, **P < 0.01, ***P < 0.001).
-
The C-terminus of core protein contains four blocks of arginine residues, which play an important role in viral DNA synthesis and genome packing (Yu and Summers 1991; Hatton et al. 1992). Therefore, we constructed four mutants, each involving one of the four blocks of arginine residues (Fig. 5A). Western blot results confirmed that each mutant was expressed normally (Fig. 5B). In the immunoprecipitation experiment, core protein mutants were tagged using HA, and they were designated M1-HA, M2-HA, M3-HA and M4-HA. FLNB-GFP or GFP vector were co transfected with the Core-HA plasmid or one of the mutant plasmid into HEK 293T cells. This immunoprecipitation result demonstrated that FLNB-GFP could pull down Core-HA, but it not the mutants (Fig. 5C). The result indicates that a mutation in any of the arginine residue blocks can prevent the interaction between filamin B and core protein. Based on this result, we can preliminarily conclude that the four blocks of arginine residues in core protein are potential interaction sites.
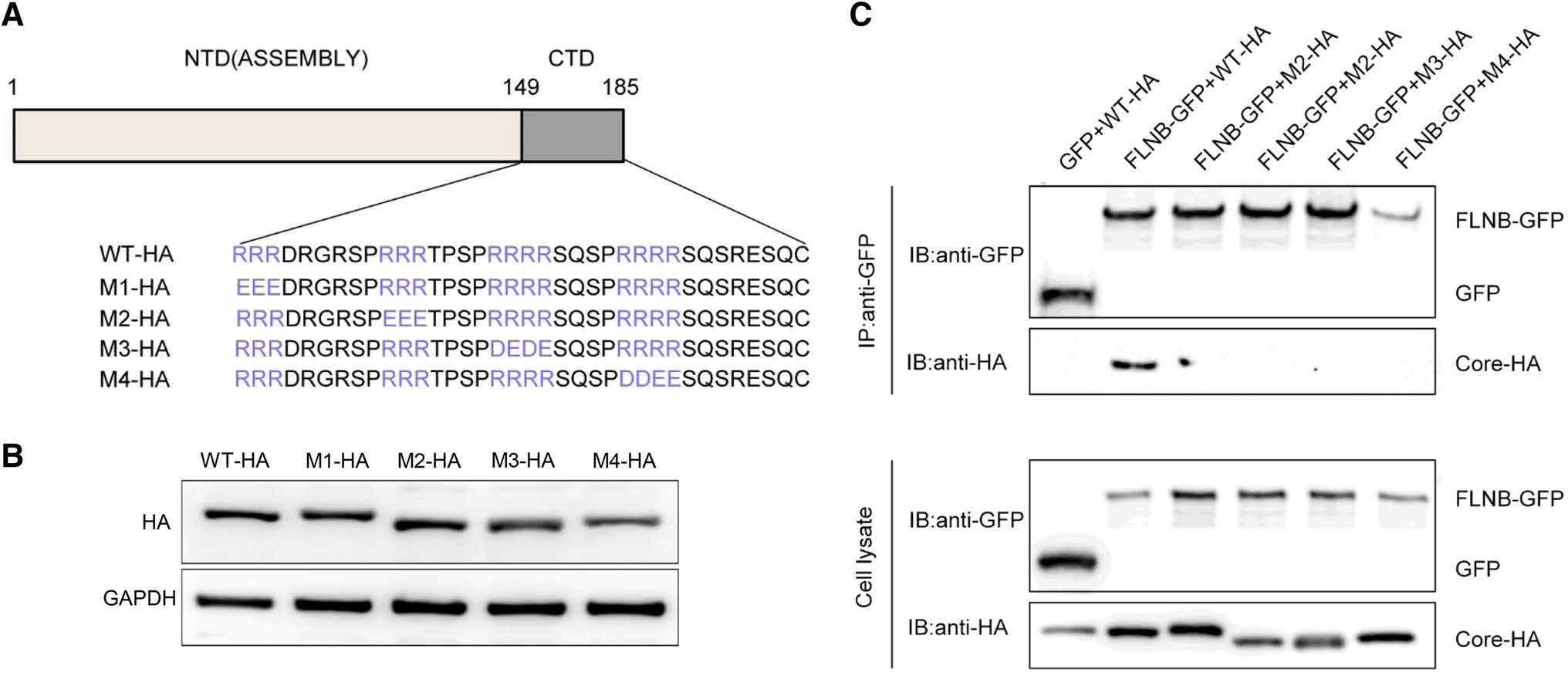
Figure 5. Potential interaction sites in core protein are shown. (A) Schematic presentation of Core-HA mutants. (B) Western blot analysis was performed to detect the expression of the mutants. (C) Co-immunoprecipitation assay was performed to confirm the interaction between filamin B and core protein mutants. A filamin B plasmid was co-transfected with a wild-type or mutated core protein plasmid into HEK 293T cells. Filamin B was immunoprecipitated using GFP. Co-precipitated core protein was detected by immunoblotting with anti-HA antibodies (top). Western blot analysis of the coprecipitated filamin B and GFP is shown using anti-GFP antibodies (top). Cell lysates before immunoprecipitation were probed with antiGFP antibodies (bottom).
Filamin B Interacts with Core Protein in Different Cell Lines
Filamin B Colocalizes with Core Protein
Interaction between Filamin B and Core Protein Promotes HBV Replication
Filamin B Knockdown Inhibits the Replication of HBV
Potential Interaction Sites in Core Protein Are Identified
-
Filamin B, a cytoskeleton protein, serves as an important molecular scaffold for the connection and coordination of various cellular processes (Stossel et al. 2001). It has been reported that the C-terminal region of filamin B interacts with HBV core protein. In addition, core protein plays an important role in the HBV life cycle (Hatton et al. 1992), which necessitates that the function of the interaction between filamin B and HBV core protein be clarified. Our findings confirm filamin B is an important host factor in the HBV life cycle, and strongly indicate that the interaction between filamin B and core protein plays an important role in regulating HBV replication, which provide new insight into the relationship between HBV and host factors.
To confirm the interaction between filamin B and HBV core protein, we also used Core-HA to pull down FLNBGFP to examine the specific interaction between them, but the immunoprecipitation experiment failed. We propose that the molecular weight of FLNB-GFP (305 kD) and core protein (25 kD) might be too different, making it difficult for the small core protein to pull down the large FLNBGFP. Zhao et al. investigated the specific interaction between the host factor ceruloplasmin (CP, 130 kD) and HBV middle surface protein (MHB, 35 kD), and they had similar problems with the reverse immunoprecipitation experiment, which indicated that MHB could not pull down CP (Zhao et al. 2017). Based on our results and the results from the other lab, it is hypothesized that molecular weight may affect the pull-down result during immunoprecipitation experiments, which should be investigated in a further study.
In this study, we found that the co-transfection groups (FLNB-GFP, Core-HA group) expressed a higher level of core protein than the control group (GFP, Core-HA group). We propose that filamin B may be involved in modulating capsid assembly and stability and the expression of core protein. As reported by Liu et al., the interaction between Dicer-2 (DCR-2) and Caenorhabditis elegans RNAi protein RDE-4 homolog (R2D2) could stabilize both proteins in Drosophila S2 cells (Liu et al. 2003). Therefore, it is reasonable to hypothesize that filamin B promotes the stability of core protien. Through further efforts are needed to clarify this conclusion. This study showed that the interaction between filamin B and core protein promotes HBV replication. However, whether filamin B, core protein, or both influence each other during this process is still unknown. However, we know that the ubiquitination of filamin B is different between a normal hepatoma cell line and an HBV-expressing hepatoma cell line. Thus, core protein may regulate HBV replication by influencing filamin B.
Filamin A is similar to filamin B, and it can interact with HIV-1 Gag protein. Subsequently, the interaction could contribute to viral particle assembly (Cooper et al. 2011). Gag protein is an essential HIV component in terms of HIV assembly and release (Wills and Craven 1991; Freed 1998), which is similar to the function of core protein in HBV life cycle. Therefore, filamin B may contribute to the assembly of core protein into a nucleocapsid.
As a pleiotropic keystone in the HBV life cycle, core protein can self-assemble to form the viral capsid, which can be targeted by antivirals (Zlotnick et al. 2015). NVR3-778, a new drug targeting the HBV capsid, has high antiviral activity in mice with humanized livers suffering from stable HBV infection (Klumpp et al. 2017). Similarly, filamin B is a potential drug target, as targeting it would disrupt the formation of the HBV capsid.
In conclusion, this study further validated the interaction between filamin B and HBV core protein, suggesting that filamin B may be an important host factor that targets core protein, which provides a new insight to understand the interaction between HBV and host factors. In addition, due to the properties of the interaction between filamin B and HBV core protein, filamin B is a potential drug target.
-
We thank Prof. Arnoud Sonnenberg (Division of Cell Biology, Netherlands Cancer Institute) for his kind gift of the plasmid Filamin B -GFP. We would also like to thank Prof. Xinwen Chen (State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China) for his kind gift of the plasmid Core -HA. This research was supported by the Postdoctoral Science Foundation of China.
-
JFM, DYG and LC conceived the study and participated in the design. YLL and YSS performed the experiments and drafted the manuscript. FYS, RH and CLL analyzed data. JFM and DYG finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: