-
Dear Editor
Marine mammals are widely distributed and can be found almost in all coastal waters and coastlines around the world. The interface areas between marine and terrestrial environments provide natural habitats for aquatic and semiaquatic mammals as well as for reservoir species of avian influenza viruses (AIV) (Runstadler et al. 2013). Previous studies showed that wild aquatic birds, the natural reservoir of AIV, are able to transmit the virus to various mammals, including seals, swine, horses, muskrats, and humans (Webster et al. 1992; Reperant et al. 2009; Gulyaeva et al. 2017). Close contacts between sea mammals and wild birds on breeding-grounds could promote both interspecies transmission of AIV and virus establishment in a new host (Fereidouni et al. 2014). Various AIV subtypes (A/seal/Massachusetts/80(H7N7), A/Seal/MA/133/82(H4N5), A/Seal/MA/3807/91(H4N6), A/Seal/MA/3911/92(H3N3), A/harbour seal/Mass/1/2011(H3N8) and A/harbor seal/NL/PV14-221_ThS/2015(H10N7) etc.) have been isolated from different species of marine mammals during the last 30 years. AIV isolated from marine mammals and wild birds are closely related, which suggests that wild birds are the major source of AIV infection (Fereidouni et al. 2014; Bodewes et al. 2015). In addition, AIV can cross species barrier and replicate well in experimental mammals without prior adaptation (Driskell et al. 2012).
Caspian seal (Phoca (Pusa) capsica, Gmelin, 1788) is an endemic species, sharing habitat with migratory birds in the Caspian Sea region where migratory pathways from Africa and Asia to Siberia intersect (Danilenko and Soldatov 2015). Antibodies against H3N2 viruses circulating in the human population from 1979 to 1981 were previously found in Caspian seal sera, suggesting this virus had been introduced into Caspian seal population directly from humans (Ohishi et al. 2002). Recent studies have further shown that seals can be infected by both human and avian influenza viruses, as seals have both α-2, 6-SA and α-2, 3-SA receptors in different tissues including respiratory tract (Runstadler et al. 2013). Therefore, like swine, seals may also act as "mixing vessels" for human and avian influenza viruses, highlighting the potential of seals to create novel reassortant influenza strains that could infect mammals and even a human (Runstadler et al. 2013).
In the present study, 27 Caspian seals of 18–20 months age (17 female and 10 male) were captured during an integrated research on AIV at Zhemchuzhny island (Astrakhan Region, Russia) in 2012. Sera samples were collected from the captured seals and tested for H1, H3, H5 and H7 specific antibodies. All animals were sera negative for these subtypes. Nasopharyngeal swabs were collected and tested for the presence of AIV using real-time PCR with previously published primers and probes targeting the matrix (M) gene (Swayne et al. 1998). One PCR positive sample was collected from the eighteen months old female seal without clinical signs. The sample was inoculated into 10-day-old embryonated chicken eggs. Allantoic fluid was collected 3 days post infection (dpi). The resulting strain belongs to the low pathogenic AIV and was named as A/Caspian seal/Russia/T1/2012(H4N6) (abbreviated as A(H4N6)T1 thereafter).
The full-length genome of the isolate was sequenced using MiSeq (Illumina, USA) according to the manufacturer's instructions. Phylogenetic analysis of all viral genes was performed using the MEGA v6.0 software with the maximum-likelihood method and 1000 bootstrap replications. Phylogenetic analysis showed that all genes of A(H4N6)T1 were closely related to avian-derived influenza viruses of the classical Eurasian lineage circulating in wild birds (Table 1). In the HA gene tree, A(H4N6)T1 was clustered together with avian H4N2, H4N3, H4N6, H4N8 and H4N9 viruses (Fig. 1A). The NA gene of A(H4N6)T1 clustered within an independent lineage with containing H4N6, H7N6, H3N6 and H10N6 AIV (Fig. 1B). Previous studies show that AIV of the H4N6 subtype had been isolated from mammals. In 1999, H 4N6 viruses isolated from swine in Ontario were closely related to AIV from birds (Karasin et al. 2000). H4N6 viruses were also isolated from semi-aquatic mammals (Runstadler et al. 2013). In both cases, lungs from infected pigs and seals showed pneumonia. Probably, the infection occurred after the likely species jump from avian to mammalian species, these H4N6 viruses were probably transmitted from pig-topig whereas information on host-virus adaptation is lacking for seals. Therefore, AIV of the H4N6 subtype have the potential to pose a serious health threat to mammals, including the endangered Caspian seal.

Table 1. Genetic identities of A/Caspian seal/Russia/T1/2012(H4N6) to other avian influenza viruses.
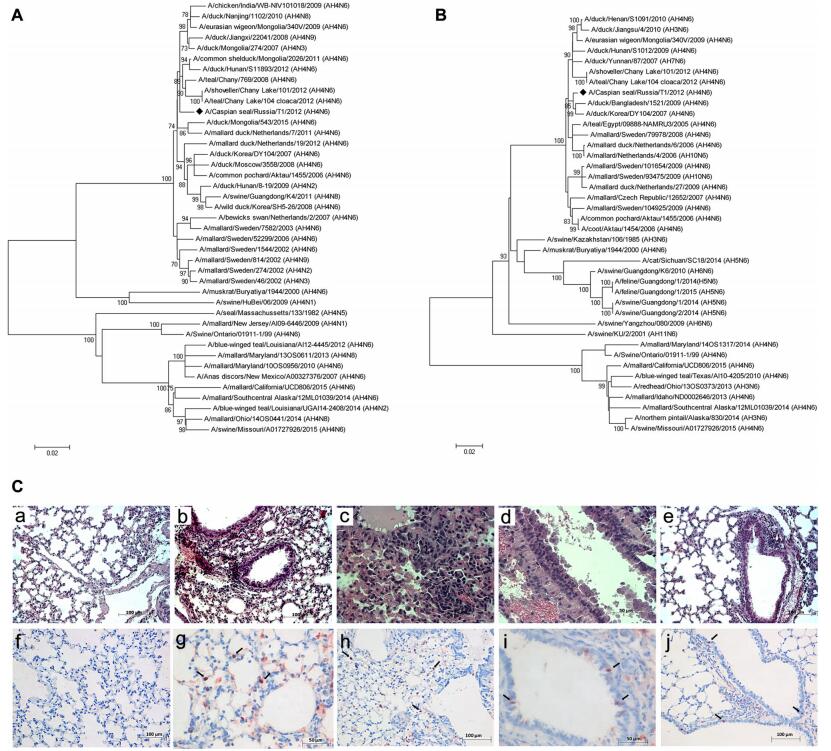
Figure 1. Phylogenetic trees of the H4 (A) and N6 (B) genes. Phylogenetical analysis of H4 and N6 genes by the maximum-likelihood (ML) method using MEGA v6.0. Bootstrap support values were generated using 1000 rapid bootstrap replicates. A/Caspian seal/ Russia/T1/2012(H4N6) is highlighted using a bold lozenge. C HEstained and immunohistochemical stained sections. C-a Intact mouse lungs. C-b Massive leucocyte infiltration in interstitial tissue 3 dpi. C-c Acute massive alveolar edema and leucocyte infiltration 5 dpi. C-d Epithelial desquamation in bronchioles, expansion of interstitial edema 7 dpi. C-e Cleared alveolar sacs 10 dpi. C-f–C-j Positive stained nucleuses in alveolocytes and epithelium cells (showed by the arrow): f-Intact mouse lungs, g-3 dpi, h-5 dpi, i-7 dpi, j-10 dpi.
Molecular characterization analysis revealed that the virus possessed several amino acids that may increase virus pathogenicity for mammals. For example, some potential pathogenic markers were found in the M1 (30D and 215A) and NS1 (42S, 41 K and 38R) proteins of A(H4N6)T1 and other mammal derived H4 strains, which could increase virulence and lethality for mice (Fan et al. 2009; Jiao et al. 2008). However, the mammalian adaptation mutations were not found in the PB2 protein (E158G, A199S, D253 N, D256G, T271A, M535T, A588I, GQ590/591SR, Q591 K, E627 K, L636F, A684S, D701 N, K702R, S714 N) (Mänz et al. 2013).
To further assess the pathogenicity of the seal-derived virus in mammals, thirty 6-to-8-week-old BALB/c mice were intranasally infected with A(H4N6)T1 (10 MID50/50μL) and twenty mice were used as controls. Clinical signs including decreased physical activity and body weight losses (~ 15% of average initial body weight) were observed 3 dpi in the infected animals. On 3, 5, 7, and 10 dpi, 5 mice per group were humanely euthanized and lung samples were collected aseptically for histopathological test, RT-PCR, and virus titration as described previously (Ilyicheva et al. 2011). Examination of the hematoxylin and eosin (HE)-stained sections of lungs of the infected mice revealed massive leucocyte infiltration and epithelial desquamation in bronchioles indicating damages in the respiratory area of lungs 3 dpi comparing with control (Figs. 1C-a, 1C-b). In addition, inflammation in the lung respiratory area intensified from 5 to 7 dpi with massive infiltration and local hemorrhages in the interstitial tissue and a pulmonary edema (Figs. 1C-c, 1C-d). Lumens of respiratory areas of lungs looked cleared from detritus at 10 dpi, but signs of interstitial pneumonia were still observed (Fig. 1C-e). Previous studies have shown that all influenza viruses that cause infection in mammals could infect epithelial cells of the upper airways and bronchioles. Highly virulent strains can also induce the pathological changes mentioned above and further cause more extensive damages of the respiratory tract and inflammation in alveoli (Guarner and Falcón-Escobedo 2009). Immunohistochemistry (IHC) staining results revealed positive staining in the lung tissues as previous described (Kovner et al. 2013). The antigen (nucleoprotein) was detected in alveolocytes and bronchioles epithelium cells nucleuses from 3 to 10 dpi (Figs. 1C-g–j). In addition, virus titers were detected in lungs of the infected mice on 3 dpi (1.75 ± 0.5 TCID50/mL), 5 dpi (3.43 ± 1.2 TCID50/mL), and 7 dpi (2.0 ± 0.4 TCID50/mL) with a peak on 5 dpi. Specific antibodies against A(H4N6)T1 were tested on 21 dpi by the hemagglutination inhibition (HI) assay as previously reported (Ilyicheva et al. 2011) and the results revealed that the sera collected from the ten infected mice displayed significant HI titers (≥ 1/80), which were not observed in the control group.
The infection caused by AIV is a serious concern worldwide, primarily due to the potential to cross species barrier to infect mammals including humans. The emergence of potentially epidemic or even pandemic strain from avian hosts would require previous adaptation to mammals (Reperant et al. 2009). In this study, we identified the H4N6 virus A(H4N6)T1, isolated from Caspian seal. The A(H4N6)T1 virus probably originated from birds. Mice study showed that A(H4N6)T1 could replicate in lungs and cause severe disease in the experimental mice without prior adaptation. The role of sea mammals as intermediate hosts or carriers of potential zoonotic pathogens remains poorly understood (Fereidouni et al. 2014), highlighting a necessity for long-term and extensive surveillance of influenza virus in marine.
-
This study was supported by RFBR (research project No.17-04-01919), the National Key Research and Development Project of China (2016YFE0205800), the National Science and Technology Major Project (2016ZX10004222), intramural special grants for influenza virus research from the Chinese Academy of Sciences (KJZD-EW-L15). YB is supported by the Youth Innovation Promotion Association of Chinese Academy of Sciences (CAS) (2017122). WS was supported by the Taishan Scholars program of Shandong Province (ts201511056). The authors thank Thijs Kuiken and Peter van Run from the Department of Viroscience, Erasmus MC for assistance in the IHC analysis, and Vladimir Petrov from the Federal Research Center of Fundamental and Translational Medicine for proofreading the manuscript.
HTML
Acknowledgements
-
The authors declare that they have no conflict of interest.
-
The work was performed in accordance with the ethical standards of laboratory animal treatment (Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes), the International Guiding Principles for Biomedical Research Involving Animals (1985), ethical norms for handling animals approved by the Biomedical Ethical Committee of the Federal Research Center for Fundamental and Translational Medicine (No. 25 of 19.11.2012) and The Rules of Laboratory Practice in the Russian Federation (Order of the Ministry of Health of the Russian Federation No.267 of 19.06.2003).














 DownLoad:
DownLoad: