HTML
-
Single-stranded (ss) DNA viruses (such as geminiviruses) are among the most successful groups of plant pathogens (Hanley-Bowdoin et al. 2013). Mastreviruses and begomoviruses in Geminiviridae are transmitted by leafhoppers and whiteflies, respectively, and can infect monocots and dicots (Hanley-Bowdoin et al. 2013). Aphid-borne Banana bunchy top virus (BBTV), an ssDNA virus belonging to the genus Babuvirus in the family Nanoviridae, naturally infects only species in the Musaceae family and is a severe threat to commercial banana production (Wu and Su 1990).
The BBTV genome is made up of at least six circular ssDNA components, all of which exhibit rolling-circle replication (Burns et al. 1995). BBTV genomic DNA components are displayed as a single open reading frame (ORF) and a 5′ intergenic region including the origin of rolling-circle replication, similar to geminiviral components, exemplified by DNA component 4 (Burns et al. 1995). The BBTV DNA 4 protein can functionally restore the movement of the CMV-Fny-ΔMP mutant (Cucumber mosaic virus-Fny isolate with the deletion of MP) (Sun et al. 2002). Protein B4 is capable of redirecting DNA 6 gene products, which preferentially translocate into the nucleus, to the cell periphery (Wanitchakorn et al. 2000). Therefore, BBTV DNA 4 and 6 gene products are considered analogous to movement protein (MP) and nuclear shuttle protein (NSP), respectively (herein referred to as proteins B4 and B6). As previously reported, geminivirus NSP and MP orchestrate the ingress/egress of viral DNAs into and out of the nucleus and mobility between cells (Rojas et al. 2001; Fondong 2013). However, BBTV MP and NSP do not display detectable sequence identity/similarity to bipartite geminiviral counterparts.
Uniquely, BBTV movement protein B4 contains a single transmembrane (TM) motif adjacent to the N-terminal domain, thereby facilitating membrane insertion (Zhuang et al. 2016). The single N-terminal TM motif along with lateral charged residues is likely equivalent in function to a signal peptide sequence used to guide proteins through the secretory pathway. Thus, the functional cleavage of protein B4, i.e., the removal of the N-terminal TM domain by proteases, may be required for its activation. In the present study, we confirmed the membrane-bound subcellular localization of protein B4, which coexists in the vasculature with DNA 4 components. We further identified a B4-derived cryptic peptide whose functional release is associated with BBTV pathogenicity. The cryptic peptide appears to act as a non-canonical modifier by host (target) protein conjugation.
-
Nicotiana benthamiana and its GFP-transgenic 16c line were used. The seeds were plated on nutrient soil and imbibed for 1 week at 25 ℃ under a 16-h-light/8-h-dark cycle. The seedlings were transferred individually into vials and kept at 25 ℃ under a continuous 16-h light/8-h dark cycle until reaching the 5-leaf stage (2-week-old N. benthamiana seedlings) for agrobacterial inoculation. In addition, BBTV-infected bananas maintained under natural conditions (20-30℃ and natural lighting) were harvested from the banana field at Fujian Agriculture and Forestry University.
-
The PVX:B4 and PVX:B4GFP constructs were assembled by cloning protein B4 and B4GFP fused/inserted between Sal I and Cla I sites of the PVX vector, respectively. The B4/B4GFP mutants were amplified using primers listed in Supplementary Table S1 and the corresponding PVX:B4 and PVX:B4GFP derivatives were obtained using a Fast Mutagenesis Kit V2 (Vazyme Biotech, Nanjing, China). All constructs were introduced into Agrobacterium tumefaciens strain GV3101 and cultured in Luria-Bertani (LB) broth overnight at 28 ℃. The inocula were transferred into fresh medium (1:50 dilution) and incubated for 6-8 h. Bacteria were pelleted and re-suspended in infiltration buffer (5 g/L glucose, 10 mmol/L MgCl2, 50 mmol/L MES-KOH, pH 5.7; including 100 μmol/L acetosyringone) until an OD600 value of 1.0 was obtained. Cultures of agrobacteria were maintained at room temperature for 1-2 h prior to infiltration into the lower epidermis of 14-day-old N. benthamiana leaves using 1-mL syringes without needles. Agrobacterium with PVX alone and infiltration buffer alone were used as negative controls. Seven-to-ten days post-infiltration, leaf tissues were harvested for total protein extraction. DNA extracted from BBTV-infected banana petioles was inoculated into the stem of N. benthamiana using a syringe, using DNA from healthy bananas as a control.
-
BBTV-infected banana petioles with (symptomatic) dark green streaks were sliced into thin sections. The samples were permeabilized in PBS (pH 7.4) containing 2% (v/v) Triton X-100 and 5% (v/v) β-mercaptoethanol for 4 h at 20 ℃. They were then fixed in PBS containing 1% Triton X-100 and 4% paraformaldehyde for 4 h. Post-fixation, samples were washed with hybridization solution three times, placed into a hybridization solution containing 1% β-mercaptoethanol and 30 μmol/l DNA fluorescent probe for 5 h, washed in 1× SSC three times, and washed twice in PBS (pH 7.4). Confocal microscopy was used to evaluate the samples for the presence of BBTV. The following FAM-labeled probe was used to visualize DNA4: B4 FAM-RC-Probe, 5′-FAM__CAGAAACCATTCGAAGAATAGTTTCACC-3′.
-
Approximately 7 days post-inoculation, newly developed leaves infected by chimeric PVX were harvested. The lower epidermis of tobacco leaves was separated/peeled using fine tweezers and tobacco mesophyll cells were placed into an enzyme solution containing 1% cellulase R-10 (Yakult Honsha, Tokyo, Japan), 0.25% macerozyme (Yakult Honsha), 0.4 mol/L mannitol, 10 mmol/L CaCl2, 20 mmol/L KCl, 0.1% bovine serum albumin, and 20 mmol/L MES (pH 5.8) for 1-2 h at room temperature (with constant agitation at ~30 rpm). The lysate was filtered using fiber sheets. The filtrate was centrifuged at low speed (~100 × g) for 5 min. The green pellet was rinsed three times using W5 buffer (150 mmol/L NaCl, 125 mmol/L CaCl2, 5 mmol/L KCl, 5 mmol/L glucose, 2 mmol/L MES pH 5.7) and re-suspended in W5 buffer. Approximately 20 µL of protoplast suspension was removed and placed onto sterile glass slides for inspection by microscopy (Nikon, Tokyo, Japan).
-
The peeled BBTV petioles (BBTV-infected and healthy samples) were individually harvested and cut into small (1-2 cm) pieces. Cooled samples were macerated for several minutes in extracting buffer [0.15 mol/L Tris-Cl pH 8.0, 0.5% (w/v) NaSO3, 0.2% (W/V) PVP-K30], ground on ice for several minutes, and filtered using 100 meshes. Chloroplasts were isolated from filtrates using an extraction kit (Sigma-Aldrich, St. Louis, MO, USA), following the supplier's guidelines. Briefly, the macerates were centrifuged for 3 min at 200 × g to remove cellular debris. The supernatant was retained and centrifuged at 1000 × g for 7-10 min. This time, the supernatant was discarded and the green pellet was re-suspended in 1× CIB. The chloroplast extracts were loaded onto a discontinuous 40%-80% Percoll gradient. After centrifugation, the green layers containing chloroplasts were harvested to isolate thylakoid membranes.
To purify the thylakoid membrane, the chloroplast suspensions were subjected to ultrasonic treatment at 4 ℃. The homogenate was centrifuged at 10, 000 × g for 10-20 min. The green supernatants were loaded onto 15%-40% sucrose gradients and centrifuged in a swinging bucket rotor (type 32, Beckman) at 4 ℃ for 4 h at 110, 000g. The separated fractions were retained for SDS-PAGE and western blot analyses.
-
Tobacco stems were cut into 0.5 cm2 sections and fixed for 2 h in PBS (pH 7.2) containing 4% paraformaldehyde and 0.1% glutaraldehyde at room temperature. After fixation, the samples were washed three times using PBS. The samples were dehydrated through an ethanol gradient of 50%, 70%, 90%, and 100% for 1 h at -20℃. Post-dehydration, mixtures of the embedding agent LR-Gold and ethanol at 1:1 and 2:1 ratios were used to permeabilize the samples for 1 h at -20℃. The samples were embedded in complete LR-Gold for 1 h at -20℃. Samples were then transferred to LR-Gold containing 0.1% BENZIL for 3 h at -20℃. Finally, the samples were placed into thin-walled tubes with LR-Gold containing BENZIL and polymerization was achieved under UV light at 20 ℃ over a 7-day period. For immuno-electron microscopy, ultra-thin sections were blocked using 2% BSA for 30 min prior to labeling with the anti-GFP antibody and 10-nm gold particles conjugated to goat antibodies against rabbit IgG (GAR10; British Bifocals International, Cardiff, UK). The specificity of labeling was monitored by incubating non-infected tobacco samples with anti-GFP IgG.
-
B4GFP fluorescence was detected using a confocal laser microscope Zeiss LSM 710 (Carl Zeiss, Oberkochen, Germany) equipped with a 40× C-Apochromat objective. GFP expressed in tobacco leaves was detected by long wave UV excitation at 488 nm and emission at 509 nm. Ultrathin sections of banana petioles and immuno-gold particles were observed by transmission electron microscopy (Hitachi H-7650).
The banana samples were harvested and cut into thin (0.2 cm2) slices and fixed in PBS (pH 7.2) containing 4% paraformaldehyde, 0.1% glutaraldehyde, 0.1% Triton X-100, and 2% β-mercaptoethanol at 26 ℃ for several hours. After fixation, the samples were dehydrated using a gradient of ethanol and then coated in embedding agent at 26 ℃ for several hours. The polymerizing patches were prepared for ultrathin slicing and stained in 1% uranyl acetate prior to observation by TEM.
-
Approximately 20 g of 'green streak' sections from BBTV-infected banana petioles were peeled, flash frozen in liquid nitrogen, and ground into a fine powder. PBS pH 7.4 [containing 0.5% (w/v) NaSO3, 0.2% (W/V) PVP-30, and complete protease inhibitor (Roche, Basel, Switzerland)] was added to the sample prior to vortexing and homogenization in an ice bath. The sample was passed through a 100-mesh filter and subsequently centrifuged once at 10, 000 × g for 30 min and again at 40, 000 × g for 20 min at 4 ℃. The remaining supernatant was centrifuged at 150, 000 × g for 4.5 h at 4℃. The pellet was re-suspended in 500 µL of 50 mmol/L PBS, pH 7.4. The suspension was aliquoted (50 µL per vial) for co-immunoprecipitation (CoIP), and stored at -70℃. Total proteins from healthy bananas were extracted following the above procedure.
-
Approximately 0.5 g of PVX:B4GFP-infected N. benthamiana (GFP-expressing N. benthamiana as a control) leaves were homogenized using lysis buffer [50 mmol/L Tris-HCl pH 7.5, 5 mmol/L MgCl2, 150 mmol/L NaCl, 0.1% Tween 20, 5% (v/v) β-mercaptoethanol, and a protease inhibitor cocktail (Roche)] on ice. Lysates were centrifuged twice at 30, 000 × g at 4 ℃ for 20 min. Approximately 40 µL of supernatant was transferred to a 0.5-mL tube and 10 µL of 5× SDS sample buffer (CW Biotech) was added. The remaining supernatant was incubated with 20 µg of anti-GFP antibody at 16 ℃ for 0.5 h and then transferred to another column pre-filled with protein A/G agarose beads (Abmart). Next, the beads were rinsed thoroughly using a 6-fold column volume of the lysis buffer and 50 μL of 5× SDS sample buffer was used to suspend the beads after the washing step. Subsequently, all samples were boiled at 98 ℃ for 5 min to disrupt disulfide bonds and linearize the peptides. The treated protein samples were centrifuged at 14, 000 × g for 5-10 min, separated by 12%-15% SDS-PAGE, and transferred onto a PVDF membrane. Immunoblotting (IB) was performed using anti-B4 and anti-GFP antibodies (Genscript). Specifically, 1× TBST (50 mmol/L Tris-Cl pH 8.0, 150 mmol/L NaCl, 0.05% Tween 20) supplemented with 5 mmol/L MgCl2 was used for anti-B4 immunoblotting at 16 ℃.
-
Approximately 0.5 g of PVX:B4GFP-infected N. benthamiana (GFP-expressing N. benthamiana as a control) leaves was homogenized using lysis buffer [50 mmol/L Tris-HCl pH 7.5, 5 mmol/L MgCl2, 150 mmol/L NaCl, 0.1% Tween 20, 5% (v/v) β-mercaptoethanol, and a protease inhibitor cocktail (Roche)] on ice. Lysates were centrifuged twice at 30, 000 × g at 4 ℃ for 20 min. Approximately 40 µL of supernatant was transferred to a 0.5-mL tube and 10 µL of 5× SDS sample buffer (CW Biotech, Beijing, China) was added. The remaining supernatant was incubated with 20 µg of anti-GFP antibody at 16 ℃ for 0.5 h and then transferred to another column pre-filled with protein A/G agarose beads (Abmart, Shanghai, China). Next, the beads were rinsed thoroughly using a 6-fold column volume of the lysis buffer. Approximately 50 μL of 5× SDS sample buffer was used to suspend the beads after the washing step. Subsequently, all samples were boiled at 98 ℃ for 5 min to disrupt disulfide bonds and linearize the peptides. The treated protein samples were centrifuged at 14, 000 × g for 5-10 min, separated by 12%-15% SDS-PAGE, and transferred onto a PVDF membrane. Immunoblotting (IB) experiments were respectively performed using anti-B4 and anti-GFP antibodies (GenScript, Piscataway, NJ, USA). Specifically, 1× TBST (50 mmol/L Tris-Cl pH 8.0, 150 mmol/L NaCl, 0.05% Tween 20) supplemented with 5 mmol/L MgCl2 was used for anti-B4 immunoblotting at 16 ℃.
To capture the target protein fragments associated with the B4-derived cryptic peptide in infected banana, the total banana protein extracts separated by centrifugation at 40, 000 × g and the supernatant were used for CoIP at 16 ℃. Similarly, CoIP using anti-B4 IgG required the addition of appropriate amounts of Mg2+ into protein suspensions to a final concentration of 1.0 mmol/LM. Next, 20-30 µg of anti-B4 IgG was sequentially added and incubated at 16℃. The mixture was aspirated into columns pre-filled with protein A/G agarose beads (Abmart). The beads were gently rinsed by running buffer through the column and re-suspended in 5× SDS sample buffer.
-
Protein bands were stained using Coomassie brilliant blue (CBB) R-250 staining and excised from gels for further analysis. The Coomassie-stained gel slices of target proteins were excised and applied to a 96-well plate, de-stained twice in 200 µL of 15 mmol/L potassium ferricyanide and 50 mmol/L sodium thiosulfate (1:1) and then dried twice with 200 μL of acetonitrile. The dried gels were incubated in a pre-chilled digestion solution (trypsin 12.5 ng/µL and 20 mmol/L NH4HCO3) for 20 min and incubated overnight at 37 ℃. Finally, the fragmented peptides were pelleted using an extraction solution (5% formic acid in 50% acetonitrile) and desiccated under a stream of N2.
The dried peptides were dissolved in solvent A (5% acetonitrile, 0.1% formic acid) and analyzed using a Triple-TOF 5600 system (AB SCIEX, Framingham, MA, USA). Briefly, peptides were separated on a reverse-phase column (ZORBAX 300SB-C18 column, 5 μm, 300 Å, 0.1 × 15 mm; MicroMass, Cary, MA, USA) using an Eksigent 1D PLUS system (AB SCIEX) at an analytical flow rate of 300 nL/min. The peptides were separated with a linear gradient from 5% to 40% of solvent B (0.1% formic acid/90% acetonitrile) over 2 h. Survey scans were acquired from 800 to 2500 Da with up to 15 precursors selected for MS/MS and dynamic exclusion for 20 s. For protein identification, MS/MS data were processed using MASCOT version 2.3.02 (Matrix Science, London, UK) and the Nicotiana subset of the NCBI sequence databases. Peptides with significance scores over the "identity threshold" were recorded and the sequence was confirmed by an artificial analysis according the b and y ions.
Plant Materials and Growth Conditions
Plasmid Construction and Viral Inoculations
Fluorescence in situ Hybridization (FISH) Detection of BBTV
Protoplast Preparation of N. benthamiana Mesophyll Cells Infected by PVX:B4GFP
Preparation of Thylakoid Components from BBTV Petioles
Immuno-EM Assay
Imaging and Microscopy
Total Protein Extraction from BBTV-infected Stems
Western Blotting
Co-immunoprecipitation (CoIP) assay
Mass Spectrometry Detection
-
BBTV infection usually causes typical dark green streaks in banana petioles (Fig. 1A), suggesting the gross aggregation of chloroplasts in BBTV-infected vasculature, relative to the evenly distributed light green coloration in healthy petioles (Fig. 1B). To further evaluate the association of the vasculature and chloroplasts with BBTV, the BBTV-infected petiole veins were harvested for in situ DNA hybridization. BBTV DNA4 component was detected within plant vascular tissues and adjacent chlorophyllous companion cells of the phloem during viremia (Fig. 1C) and was absent from the same tissues of healthy bananas (Fig. 1D).
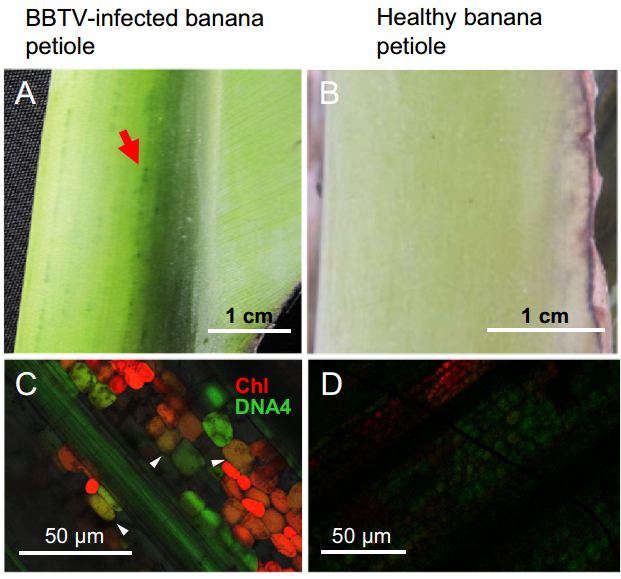
Figure 1. Banana bunchy top virus DNA4 component exists in the vasculature and companion cells. Dark green streaks (marked by a red arrow) of BBTV-infected banana petioles are clearly visible (A), whereas light green coloration is distributed evenly in the petiole of a healthy banana (B). DNA hybridization signals produced from fluorescent probes indicated that BBTV DNA4 component accumulates in the vasculature and chlorophyllous companion cells (denoted by white arrow heads) (C). No hybridization signals were detected in healthy banana cells; only chlorophyll was detected (D).
-
To determine the molecular characteristics of B4 protein, we first traced its definitive subcellular localization using a functionally intact B4GFP conjugate. We selected protein B4 from the BBTV Hainan isolate, which consists of 117 amino acids ordered into three domains: an N-terminal region (1-13 aa), a TM motif (position 14 to position 36), and a C-terminal tail (Fig. 2A). Based on PVX:B4GFP construct expression in N. benthamiana via the Agrobacterium infiltration method, the green fluorescence of B4GFP after 7 days post inoculation (d.p.i.) was distributed throughout the stem, particularly in the vasculature of N. benthamiana under blue-light excitation (Supplementary Figure S1A). B4GFP was found in the cytoplasmic membrane and appeared to travel via a symplastic route to the neighboring cells/vasculature in leaf veins (Supplementary Figure S1B). Furthermore, B4GFP accumulation in foci in the apoplastic region of the stem was identified by immunogold labelling (Supplementary Figure S1C, S1D).
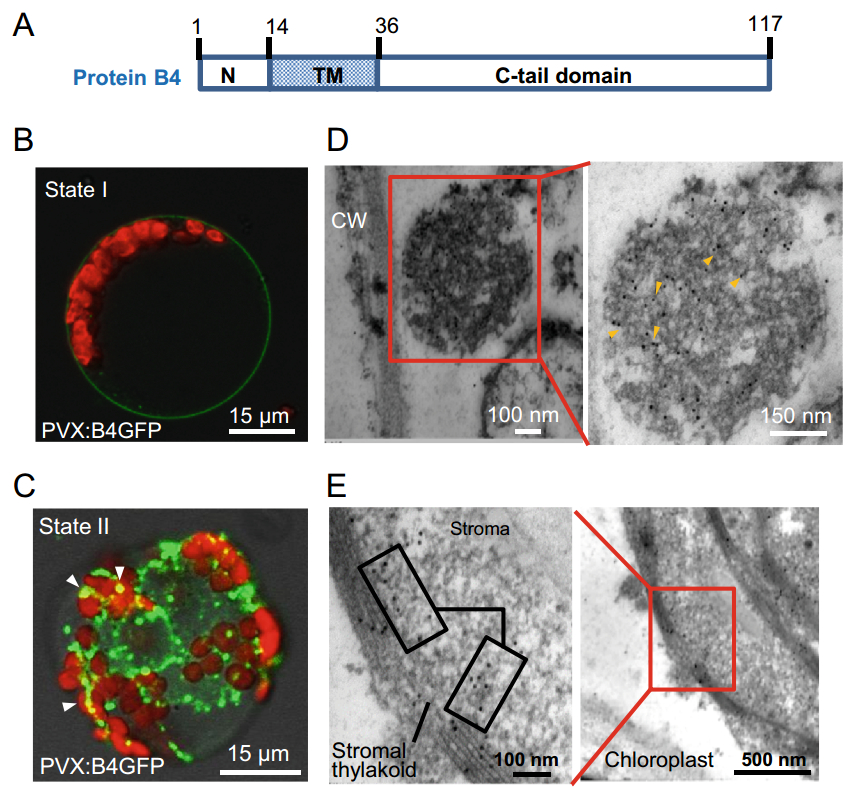
Figure 2. Subcellular localization of Banana bunchy top virus protein B4 fused to GFP. A Schematic representation of protein B4 domains from the BBTV Hainan isolate. Positions 1–13 make-up the N-terminal domain; positions 14–36 form the transmembrane domain (TM), and the remaining 37–117 C-tail domain. B B4GFP was located in the periphery of protoplasts prepared from PVX:B4GFPinfected Nicotiana benthamiana in expression state I. C B4GFP moves into the intracellular reticula and chloroplasts in granular form in expression state II. D The granular compartment adjacent to the cell wall (CW) of infected leaves was labeled immunologically using 10-nm gold particles (denoted by yellow arrowheads) for B4GFP. E Immuno-labeled gold particles localized at the stroma and thylakoid membrane of B4GFP entering the chloroplast. Images represent experiments performed on at least two independent occasions.
After B4GFP entered the foliage along with vascular tissues, expression states in different mesophyll cells were visualized. First, B4GFP circled the periphery of mesophyll cells prior to entry into the cytoplasm (Fig. 2B). Once inside the cell, it mainly aggregated into granule-like structures with distinct green fluorescence; a small portion overlapped with chloroplasts (containing chlorophylls), identified by red fluorescence (Fig. 2C). To further examine the ultrastructural location of B4GFP, ultrathin sections were assayed by immunoelectron microscopy. The granular compartment composed of filaments was specifically marked by colloidal gold particles after anti-GFP immunogold labelling (Fig. 2D). B4GFP deposited in the chloroplastic thylakoid membranes and matrix from PVX:B4GFP-infected leaves was also identified by immunolabelling (Fig. 2E).
-
BBTV colonizes phloem tissues to establish infection in bananas, leading to the formation of dark green streaks (longitudinal in direction) along the vasculature of the petioles/pseudostems. Relative to the normal morphology of chloroplasts in healthy banana (Fig. 3A), the disruption of the outer membranes of chloroplasts is induced by BBTV infection (Fig. 3B). Surprisingly, virus-like particles accumulate into crystalline assemblages that are surrounded by thylakoid membranes in BBTV-infected banana petioles (Fig. 3C). Accordingly, the coat protein of BBTV was detected in thylakoid components using an anti-CP antibody (Fig. 3D).

Figure 3. The disruption of chloroplasts upon BBTV infection accompanies the crystalline aggregation of virus-like particles enveloped by the thylakoid membrane and a substantial increase of ROS. A, B Disruption of the outer membrane and vacuolization of the broken chloroplast from healthy banana, in contrast to the intact envelope of chloroplasts from healthy banana. C The thylakoid membrane fused to vesicle/organelles encompassed the crystalline-like particles. D BBTV coat protein (CP) was specifically detected in the thylakoid pellet from BBTV-infected banana. E Significantly deeper NBT staining in sieve-tube plate between neighboring cells in infected petioles indicated that ROS were robustly elevated relative to levels in healthy banana. Scale bars 1 μm (A, B), 500 nm (C), and 60 μm (E).
A dramatic increase in reactive oxygen species (ROS) levels in sections of BBTV-infected petioles was indicated by dark NBT staining compared to light dyeing in corresponding tissues of healthy bananas, especially in sieve-tube plates of vascular cells of infected bananas (Fig. 3E). Collectively, our data suggest a close relationship between chloroplasts and BBTV infectivity, replication, and dissemination.
-
To characterize protein B4, the total protein contents of agro-infiltrated N. benthamiana were initially extracted in the presence of a protease inhibitor cocktail. Anti-GFP immuno-blotting revealed that protein B4GFP was cleaved into two smaller fragments (BGF1 and BGF2) by a putative peptidase (Fig. 4A). BGF1 is ~35 kDa, highly similar to the predicted size of the C-terminal domain of protein B4 (i.e., B4C) with intact GFP (26.9 kDa). Intriguingly, immuno-blots showed several anti-GFP-positive bands above 95 kDa after B4GFP challenge. Prior to SDS-PAGE, the total proteins resolved in loading buffer containing SDS underwent disulfide bond disruption and polymer disassembly by the addition of reducing agent dithiothreitol (DTT) and heat treatment. Therefore, it is highly unlikely that these protein bands are B4GFP self-aggregates. Concurrent with the PVX:B4GFP distribution in nascent tissues, the abundance of modified protein species of higher molecular weight (HMW) continued to increase from the original site of inoculation (Fig. 4A). Conversely, the abundance of BGF1 fragments from systemic (distal) leaves was lower than that in inoculated leaves (Fig. 4A), indicative of HMW species as a result of host factor conjugation with BGF1. In addition, B4-derived peptides in PVX:B4-infected 16C (GFP-transgenic N. benthamiana line) do not target GFP to form the HMW species (Fig. 4A), implying that protein B4 targets specific host factors, rather than random substrates (such as GFP).
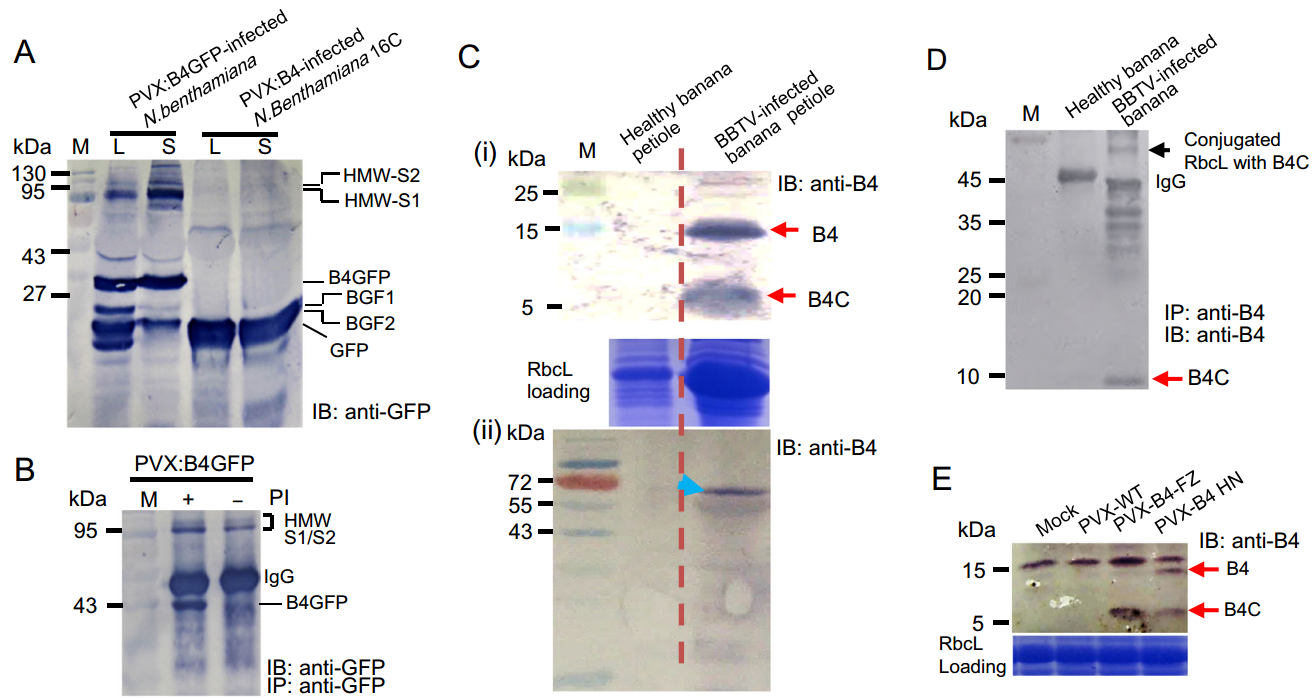
Figure 4. The discrete B4-derived functional peptide and its conjugated proteins were identified by CoIP and immunoblotting. A B4GFP and BGF1/BGF2 fragments were detected by anti-GFP immunoblotting. Higher molecular weight (HMW) species were denoted HMW-S1 and HMW-S2. B HMW species were resistant to proteolytic degradation in the absence of a protease inhibitor (PI). C Protein B4 and B4- derived cryptide (denoted B4C) were confirmed in BBTV-infected banana by anti-B4 immunoblotting. The shifted bands (denoted by an arrow) above RbcL were probed in total protein extracts of BBTVinfected banana petioles. There were no positive signals in healthy banana petioles. D The discrete B4C and B4C-conjugated RbcL species were immuno-precipitated by anti-B4 Ab and were sequentially subjected to SDS-PAGE and immunoblotting by an anti-B4 Ab. E Protein B4 and B4C also were detected in PVX:B4HN/B4FZ (from BBTV HN and BBTV FZ isolates, respectively) inocula.
We utilized CoIP to further investigate the target(s) of protein B4. The immuno-precipitates from PVX:B4GFP-infected N. benthamiana revealed a prominent HMW GFP-positive signal, which seemed to be resistant to degradation in the absence of protease inhibitors and perhaps resistant to proteasomal cleavage (Fig. 4B). The immunoprecipitated HMW species were excised from an SDS-PAGE gel, processed by trypsin, and evaluated by liquid chromatography mass spectrometry (LC-MS). Peptide mass spectra corresponded to ribulose bisphosphate carboxylase oxygenase large subunit (Rubisco LS, or RbcL), elongation factor 2 (EF2), and BGF1 fragment (Supplementary Figure S2, S3). The migration patterns of the modified proteins HMW-S1 (~90 kDa) and HMW-S2 (~132 kDa) were consistent with those of RbcL (~55 kDa) and EF2 (~95 kDa) in complexes with a single BGF1 (~34 kDa, B4C+GFP).
The full-length protein B4 and its cleaved B4C fragment were specifically probed in the protein extract from natural host banana petioles infected by BBTV (Fig. 4C-ⅰ). Similarly, an HMW anti-B4-positive signal above RbcL was specifically probed in the BBTV-infected banana protein extract by western blotting (Fig. 4C-ⅱ). We co-immunoprecipitated the HMW species in the protein extract of BBTV-infected banana using anti-B4 antibodies. The HMW species above RbcL was identified as the conjugate of RbcL and B4C by matrix-assisted laser desorption ionization time-of-flight tandem mass spectrometry (MALDI-TOF-MS/MS) (Supplementary Figure S4). In parallel, the immuno-precipitate was subjected to anti-B4 immuno-blotting. This analysis showed that these HMW species contained the RbcL-B4C conjugate, other presumptive targets, and discrete B4C peptide (Fig. 4D). B4C could be detected in the PVX:B4 inocula as well (B4HN and B4FZ individually representing protein B4 from Hainan and Fuzhou isolates) (Fig. 4E). These results provide clear evidence that the functional cleavage of protein B4 is conserved in various BBTV isolates and different plant taxa.
-
The tangible scission of protein B4 is likely executed by proteases. To identify if highly conserved arginine residues are involved in the release of the functional fragment B4C and the covalent linkage with host factors, a series of invariant positively charged arginine residues were replaced with alanine residues using site-directed mutagenesis (Fig. 5A). In contrast to the infertility induced by the PVX:B4-inoculum, PVX:B4Mut1 and PVX:B4Mut2 inocula restored flowering and seeding. All other B4 mutants retained the functionality of protein B4, leading to dwarfism and sterility (Fig. 5B). Importantly, the B4-derived peptide (B4C) was not detected in the PVX:B4GFPMut2 (RRR55-57→AAA)-inoculated plants (Fig. 5C), consistent with the absence of BGF1 from the B4GFPMut2 inoculum (Fig. 5D). Additionally, the HMW species were almost completely lacking in B4GFPMut1 (K40K43R47→AAA) and B4GFPMut6/7 (R99-100→AA) inocula, despite the presence of BGF1 (Fig. 5D). Presumably, RRR55-57 is necessary for the release of the B4-derived cryptide and other arginine residues could be responsible for conjugation to host factors. Because the mutation of protein B4 at RRR55-57 prevented cryptide release from full-length protein B4 as well as B4C (~8 kDa) and BGF1 (~34 kDa), the BGF1 cleavage site (named the P1 position) was located at RRR55-57 or adjacent residues. However, the definitive cleavage site at protein B4 needs to be confirmed, in addition to the identification of host/plant endopeptidases involved in this proteolytic processing.
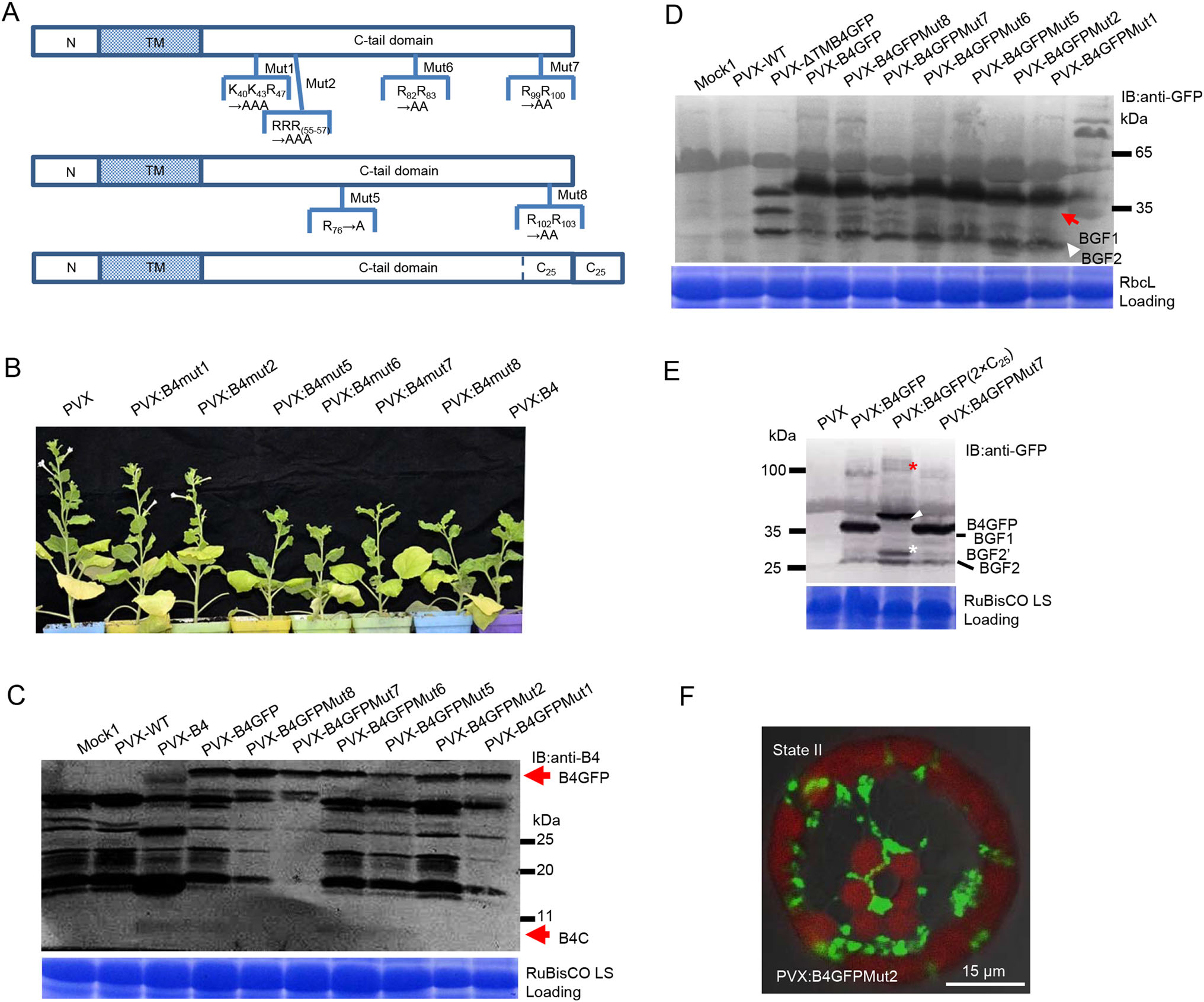
Figure 5. Crucial arginine (Arg) residues are required for the release of the B4-derived cryptide. A Schematic diagrams of mutation positions in protein B4. B BBTV B4Mut1 and B4Mut2 were not pathogenic when exposed to Nicotiana benthamiana. However, B4 mutants 5–8 displayed similar wide-type symptoms. C The B4-derived functional peptide is absent in the PVX:B4GFPMut2 inoculum. D BGF1 can be detected in PVX:B4GFP and its mutant inocula, other than the PVX:B4GFPMut2 inoculum. The anti-GFP-positive HMW species are lacking in the Mut2 inoculum, concomitant with the absence of BGF1. BGF2 and BGF1 are denoted by red and white arrowheads, respectively. E The C25 duplicate contains the two P2 cleavage sites; BGF20 fragment (denoted by a white asterisk) resulted from the incomplete proteolysis of the C25 duplicate. BGF10 is marked by a white arrowhead. The up-shifted migration of HMW species (denoted by a red asterisk) might indicate a conjugate with BGF10 containing another C25 duplicate. F GFP fluorescence surrounds the periphery of chloroplasts but does not appear inside the organelles in plants treated with B4GFPMut2.
It is likely that the P2 position in the C25 region contributes to the generation of BGF2 because BGF2′ and B4GF2 fragments were both generated from B4GFP (2×C25) fusion proteins containing two C25 repeats (Fig. 5E). BGF2′ may be a product of incomplete cleavage at the P2 position. Notably, B4GFP (2×C25) also can be processed into BGF1′ at the P1 position, which contains two C25 repeats and is larger than B4GF1. The upward shift of HMW species (denoted by a red asterisk in Fig. 5D) from the B4GFP (2×C25) inoculum was easily identifiable, unlike the changes of HMW species in B4GFP and B4GFPMut7 inocula. These observations imply that HMW species formation is mediated by conjugation with BGF1 rather than BGF2. Unlike the intact protein B4GFP, B4GFPMut2 did not overlap (co-localize) with chloroplasts but accumulated around the periphery of the organelle (Fig. 5F), thereby providing subcellular evidence for non-covalent linking to chloroplast RbcL or other proteins. Even though RbcL and EF2 are likely targets of this B4-derived modifier, these complexes does not account for the infertility of plants under B4 challenge; therefore, we predict the presence of other molecular targets or functional contribution by the liberated N-terminal fragment. It is possible that the combined activities of protein B4 and its released cryptides have indirect consequences that compromise plant-BBTV homeostasis during viral infection.
BBTV DNA4 Components Localize in Vascular Tissue and Neighboring Chlorophyllous Companion Cells
Subcellular Localization of BBTV Movement Protein B4
Chloroplast Disruption and ROS Elevation in BBTV-infected Banana Petioles
A Functional Peptide is Released from Protein B4 and Covalently Targets Host Proteins
Proteolytic Cleavage of Protein B4 Is a Pre-Requisite for Switching-on Protein Targeting Activity
-
Virus-infected plants display a common set of symptoms, such as chlorosis and mottling of the leaves, indicating that these organelles are deconstructed and damaged (Mandadi and Scholthof 2013). BBTV DNA4 components localize in vascular tissues and co-occur in companion cells where they enter chloroplasts (indicated by the merged image of the hybrid signal of the DNA 4 probe and auto-fluorescence of chlorophyll in Fig. 1D). Protein B4 enters the chloroplast as well (Fig. 2C, 2E). Furthermore, the virus-like particles are enveloped by the thylakoid membrane in BBTV-infected banana petioles (Fig. 3C). This suggests that BBTV exploits the chloroplast platform for replication and colonization. It is plausible that BBTV could directly usurp the energy produced in chloroplasts by photophosphorylation for self-advantage. Chloroplasts play pivotal roles in early immune responses, such as the production of antimicrobial ROS (de Torres Zabala et al. 2015). A homolog of bacterial respiratory Complex I, NDH-1, is involved in cyclic electron transfer around photosystem I within chloroplasts (Endo et al. 2008). NDH-1 catalyzes the substantial turnover of NADH into NAD+ accompanying the production of ROS. Therefore, a vast amount of ROS likely results from a significant elevation of NDH catalytic activity upon BBTV infection. The chloroplast essentially acts as a battling arena between host and BBTV.
BBTV movement protein B4 is a pathogenic determinant and is associated with chloroplasts. Nevertheless, intact protein B4 acts as a precursor and is processed proteolytically into the B4C fragment, which subsequently targets RbcL. Although the biological function of the conjugate of RbcL and B4-derived peptide (i.e., B4C) remains unclear, EF2, a ribosomal component that facilitates and orchestrates translational elongation, is also conjugated to B4C in PVX:B4GFP N. benthamiana inocula. Similar to the circumvention of antiviral translational suppression used by geminiviruses in plant hosts, BBTV likely intercepts cellular factors or hijacks the translational apparatus for the synthesis of viral proteins in plant organelles, such as chloroplasts.
The conjugation of the B4C fragment to other proteins seems to be similar to that of ubiquitin (Ub) in eukaryotes. Ub and Ub-like factors are regulatory proteins, functioning via their covalent attachment to substrates/targets. Usually, Ub is also produced from precursors by deubiquitinase and is further conjugated to substrates via the E1, E2, and E3 cascades (Komander and Rape 2012). Viral pathogens directly recruit the ubiquitination system by the mimicry of editing proteins or eukaryotic Ub itself (Randow and Lehner 2009; Alcaide-Loridan 2012). Host Ubs also exist in certain mammalian (e.g., poxvirus, herpes simplex virus, parainfluenza virus) and insect viruses in phospholipid-modified forms, and viral Ub-like genes in the genomes of Baculoviridae (group I nucleopolyhedrovirus) enhance pathogenicity (Guarino et al. 1995). Although sequence composition differs between the B4-derived peptide and Ub, they have similar molecular weights and are capable of conjugation with protein targets.
In addition to canonical ATP-dependent ubiquitination, NAD-dependent ubiquitin transfer to host proteins is mediated by a single bacterial effector SdeA (Bhogaraju et al., 2016; Qiu et al., 2016; Kotewicz et al., 2017). This non-canonical serine ubiquitination of target proteins via a phosphoribosyl moiety linkage with Arg42 in ubiquitin involves ADP-ribosyltransferase and phosphohydrolase activities with the aid of an NAD+ cofactor (Bhogaraju et al., 2016). The Sde-mediated phosphoribosylation of ubiquitin blocks the classical ubiquitination cascade involved in protein degradation. Similarly, protein B4 is full of conserved arginine residues and the target species conjugated with the B4-derived cryptide was resistant to proteolytic degradation. Whether the ATP-independent ligase-like bacterial effector SdeA in plants is responsible for the covalent linkage between the B4-derived peptide and plant proteins warrants further study. Additionally, the definitive linkage mode of the B4-derived modifier to host factors remains to be determined. Our data provide strong evidence that a novel modifier encoded by a plant viral protein can modulate the interactions between ssDNA viruses and plant hosts.
-
This work was supported by grants from the Natural Science Foundation of China (No. 31301641 to J.Z.) and the Program for Qualified Personnel of Taiwan Strait West Coast (No. K8812007 to L.H.X.). We are grateful to Haitao Wang for assistance with TEM, Dr. Zhixin Liu for providing the anti-B3 (coat protein) antibody, and Dr. Zhenguo Du for valuable discussions.
-
JZ conceived of the entire project. JZ and WL performed the vast majority of experiments. PS generated some plasmid constructs and performed Western blotting. TW was involved in TEM analyses. ZW provided materials, including banana samples. LX provided constructive suggestions and co-supervised the project. JZ generated or guided all Figures. JZ and CJC interpreted the data and wrote the manuscript.
-
The authors declare that they have no conflict of interest.
Conflict of interest
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
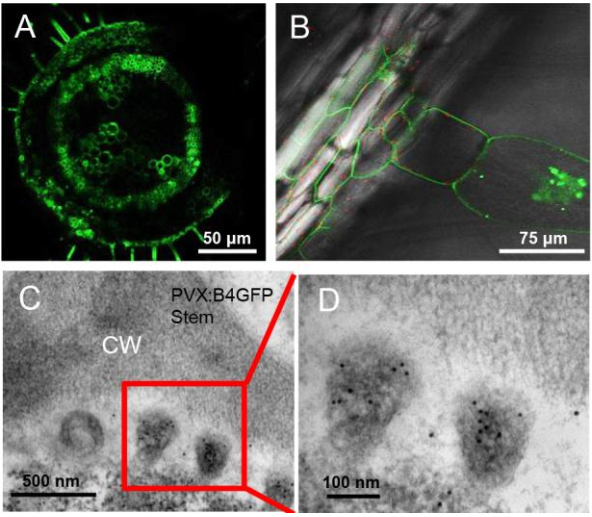
Figure S1. Cellular and subcellular localization of B4GFP in Nicotiana benthamiana. (A) The longitudinal section shows the aggregation of B4GFP in the peripheral membrane-associated region of stem cells, especially in the vasculature, in PVX: B4GFP-infected N. benthamiana. (B) B4GFP localized at the periphery of vascular bundle cells from the leaf mid-stem and entered vasculature-linking trichomes to form granules similar to those in mesophyll cells, indicating that mobile B4GFP is involved in symplastic transport. (C) Accumulation of B4GFP in vesicular tissue (stem) appeared in the apoplastic region; an enlarged image of immuno-labeled vesicles in (D) is shown. CM, cell wall.
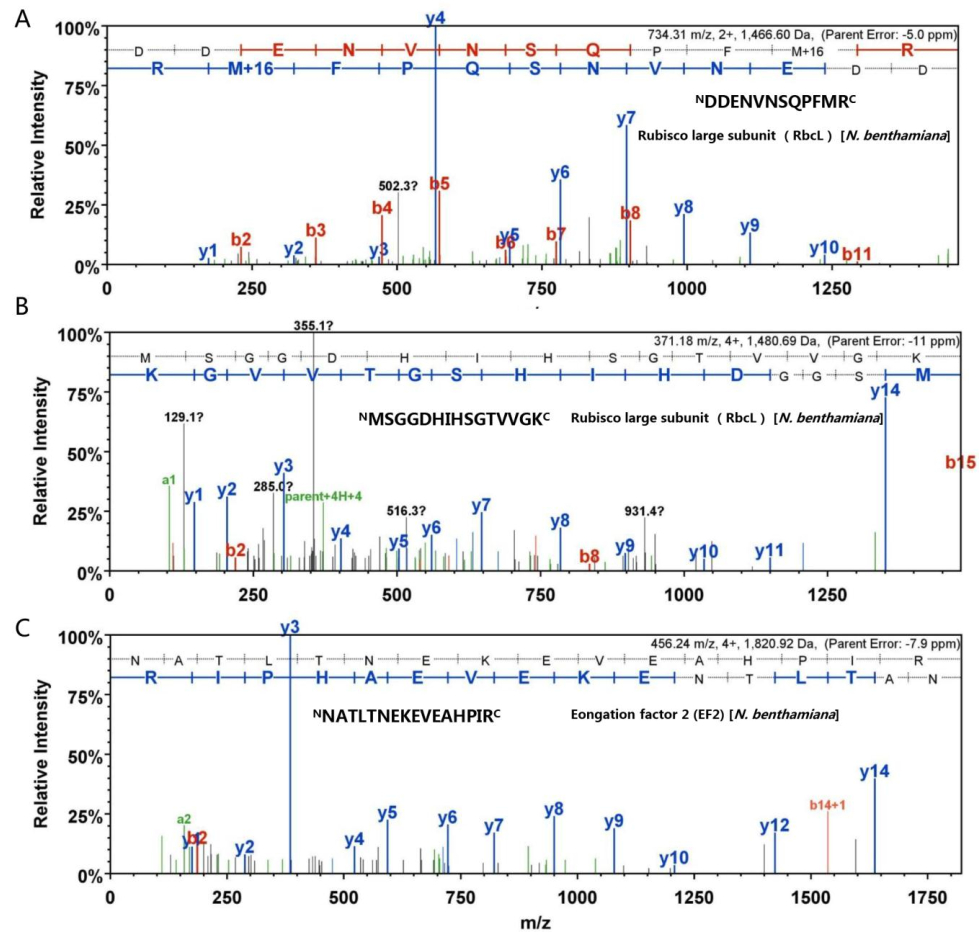
Figure S2. LC-MS spectra of peptides of RbcL and EF2 from HMW species. Three peptide sequences were determined in light of y and b ions by LC-MS. The "DDENVNSQPFMR" and "MSGGDHIHSGTVVGK" sequences were derived from RbcL (A and B). The "NATLTNEKEVEAHPIR" sequence corresponded to EF2- derived peptide (C). The individual y and b ions are sp
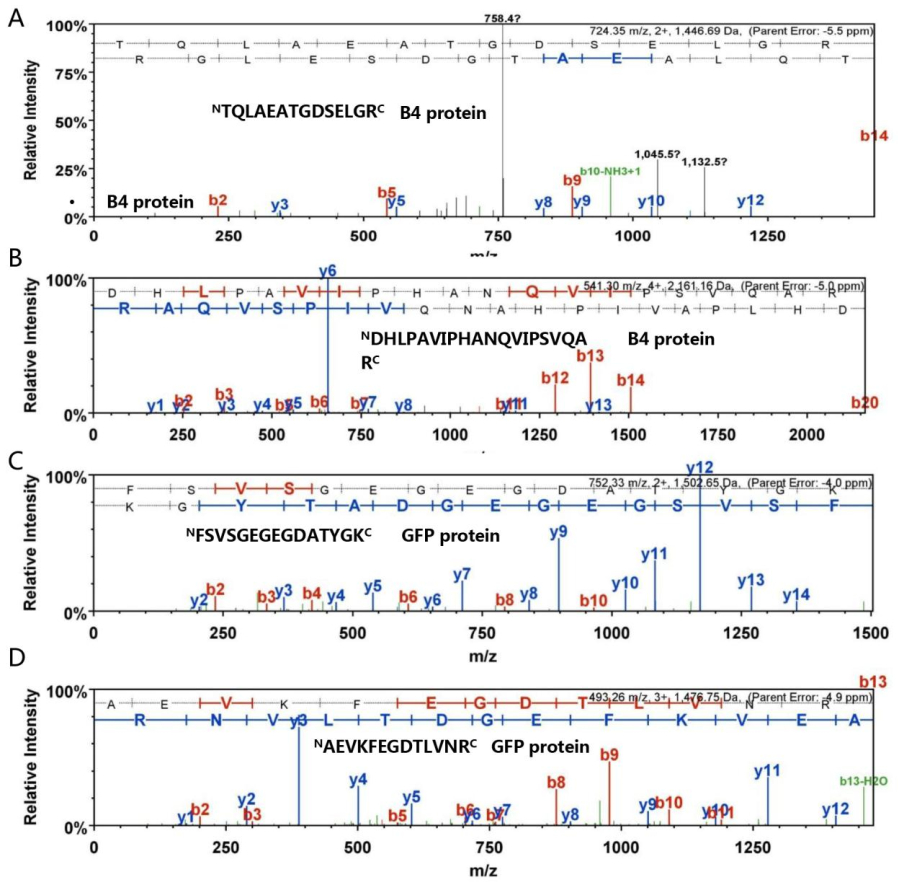
Figure S3. The LC-MS spectral data for peptides of B4 and GFP from HMW species. Three peptide sequences were determined in light of y and b ions by LC-MS. The "TQLAEATGDSELGR" and "DHLPAVIPHANQVIPSVQAR" sequences were derived from Protein B4 (A and B). "FSVSGEGEGDATYGK" and "AEVKFEGDTLVNR" sequences belonged to GFP (C and D). The individual y and b ions are specified in detail.
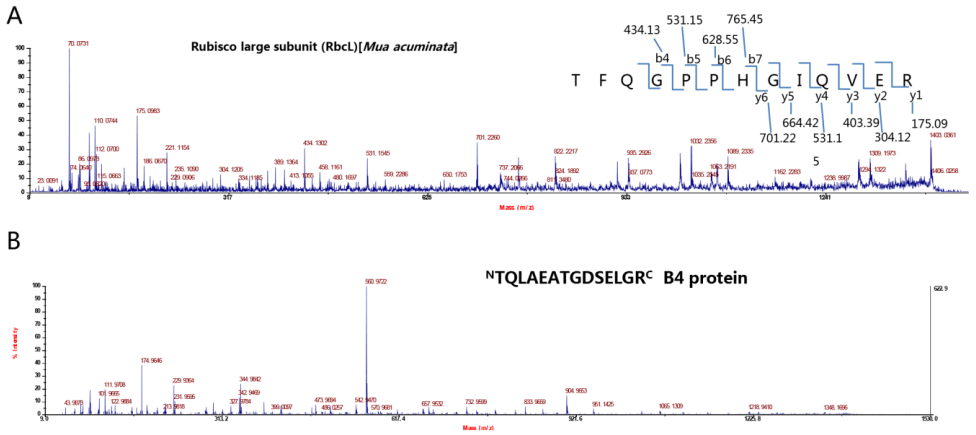
Figure S4. MS data for RbcL and B4 from HMW species in BBTV-infected immunoprecipitate. The two peptide sequences were determined according to y and b ions by MALDI-TOF/TOF MS. "TFQGPPHGIQVER" belonged to Musa RbcL (A) and "TQLAEATGDSELGR" was derived from Protein B4 (B).

Table S1. The primers for B4-derived mutants and related constructs
















 DownLoad:
DownLoad: