-
Dear Editor,
Tick-borne encephalitis virus (TBEV) belongs to the genus Flavivirus within the family Flaviviridae and includes three subtypes: Siberian, European, and Far Eastern. TBEV infects humans via tick bites; indeed, at least 10, 000 human cases of encephalitis caused by TBEV are reported annually in Russia, China, and European countries (Banzhoff et al. 2008; Carletti et al. 2017).
In China, the main subtype of TBEV is Far Eastern (Zhang et al. 2016).This subtype spread across the forest region of northeast and northwest China, with thousands of people infected annually. TBEV mainly causes severe meningitis and encephalitis. The symptoms include fever, headache, neurological disorders, and/or peripheral flaccid paralysis (Mansfield et al. 2009). Early diagnosis is crucial for the control and treatment of TBEV infection.
Currently, no commercial assay kits for TBEV infection are available in China. Hospitals most commonly use enzyme-linked immune absorbent assays and immunofluorescence assays to diagnose and monitor immune responses to TBEV infection. However, these assays are time-consuming, requiring 4 h or more for completion. Molecular techniques can complement serological assays for the diagnosis of TBEV infection. Importantly, TBEV nucleic acids can be detected within 5 days after the onset of fever. Real-time reverse-transcription polymerase chain reaction (rRT-PCR) is highly sensitive and specific. Unfortunately, because this technique requires highly specific equipment and reagents, the use of rRT-PCR is limited to well-funded and well-equipped referral laboratories. Accordingly, the use of rRT-PCR in a resource-limited setting, such as peripheral laboratories in many TBEV endemic regions in the countryside, remains limited.
In recent years, extensive efforts have been dedicated to the development of a time-saving, simple nucleic acid-based assay that relies on portable equipment for use in resource-limited settings for detection of various infectious diseases (Maffert et al. 2017). The reverse transcription recombinase polymerase amplification (RT-RPA) method, developed by Olaf Piepenburg (Piepenburg et al. 2006; Eulera et al. 2012), can be used to amplify the target gene under isothermal temperature conditions within 20 min; this method is fast, easy to use, and sensitive.
In the current study, we developed RT-RPA for TBEV. First, we selected the target sequence and encapsulated it in virus-like particles (VLPs). The target sequence for RTRPA was located in the conserved 3'-non-coding region (Senzhang strain, positions 10, 581–10, 761). VLPs containing the targeted TBEV genome fragment were constructed as template for RPA sensitivity evaluation and as a positive control for the developed RT-RPA assay. The fragment contained both the RPA target and the rRT-PCR target.
RPA primers and probes were designed according to the recommendations of TwistDX and then synthesized by Sangon Biotech (Shanghai, China). Primer and probe target specificities were evaluated using the BLAST search tool. The primers and probes matched the TBEV genome sequence but not non-TBEV sequences. In total, four forward primers and two reverse primers were generated; eight combinations of candidate primers were prepared and screened for reactivity with TBEV RNA (Supplementary Table S1). Of these primer sets, six sets (sets 1, 2, 3, 5, 7, and 8) yielded clear, specific PCR products and were identified as capable of amplifying target RNA (Supplementary Figure S1). A probe (TBEV P-RPA) was designed to accommodate all six primer sets, with 1–100, 000 copies of template RNA, and primer set 3 showed the highest sensitivity with the probe, having a limit of detection (LOD) of 10 copies/reaction (Supplementary Figure S2). Consequently, primer set 3 (TBEV-F3-RPA and TBEV-R1-RPA) was selected and used as the primer set for TBEV detection.
To test the sensitivity of the RT-RPA assay, serially diluted TBEV VLP RNA (1–107copies/μL) was used as a template in the assay. Serial dilutions of native TBEV RNA (Senzhang and Mudanjiang01 strains) were also used as a template (0.01 PFU/μL to 104 PFU/μL) in this assay. All samples were tested in duplicate with three independent runs. Probit regression analysis was performed to calculate the LOD by interpolation (Fig. 1). Based on the probit regression average, the LOD was determined to be10 copies for VLP RNA, and 0.1 PFU/reaction for the Senzhang and Mudanjiang01 strains (Fig. 1). This sensitivity was similar to those of RT-PCR assays (Schwaiger and Cassinotti 2003), which were 10 copies for VLP RNA and 0.05 PFU/reaction for the Senzhang strain (Fig. 1).

Figure 1. Sensitivity comparison of RT-RPA with rRT-PCR for TBEV detection. A Amplification curve of the RT-RPA assay determined using dilutions of VLP RNA. B Linear probit regression analysis of the RT-RPA assay determined using dilutions of VLP RNA. C Amplification curve of the RT-RPA assay determined using dilutions of Senzhang strain RNA. D Amplification curve of the RTRPA assay determined using dilutions of Mudanjiang01 strain RNA. E Linear probit regression analysis of the RT-RPA assay determined using dilutions of Senzhang and Mudanjiang01 strain RNA. The RNA template was applied to RT-RPA assays (5 μL/reaction). After incubation at 42 ℃ for 20 min in ESE Quant TS, the fluorescence intensity was measured every 20 s, and the amplification curves were generated using Tube canner studio software for the Senzhang strain, the Mudanjiang strain, and VLPs. F rRT-PCR amplification curve of the rRT-PCR assay determined using dilutions of VLP RNA. G Sensitivity of the rRT-PCR assay determined using dilutions of Senzhang strain RNA. The rRT-PCR assays were carried out in a 20-μL reaction volume containing 10 μL of One-step qRT-PCR master mix (Takara, Dalian, China), 0.4 μL each of transcriptase and polymerase, primers and probe (0.25 nmol/L each), and 2 μL tested RNA. The reaction was initiated by incubating at 42 ℃ for 5 min and then at 95 ℃ for 10 s; this was followed by 40 cycles of denaturation at 95 ℃ for 5 sand annealing at 55 ℃ for 20 s. Each sample was assayed in duplicate, and fluorescence signals were collected at 55 ℃ during every cycle. Amplification curves were generated, and the crossing point (Cp) values were calculated using Roche LightCycler 5.0
The specificity of the developed RT-RPA assay was determined using a panel of genomic RNA samples from relevant flavivirus representatives, including Japanese encephalitis, West Nile, yellow fever, Dengue (types 1–4), and Zika viruses (Fig. 2). No consistently positive signals were observed for any of these samples.
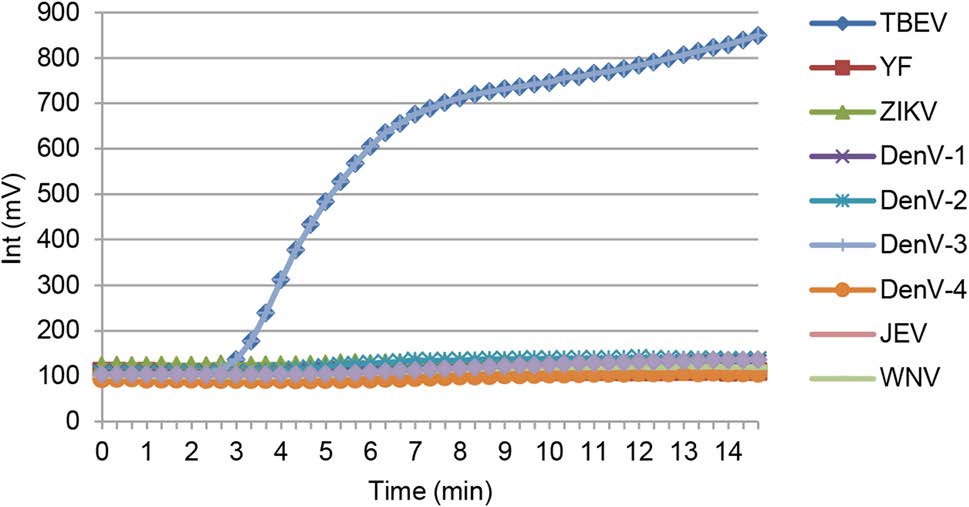
Figure 2. Specificity of the RT-RPA assay for TBEV detection. RNA samples from relevant Flavivirus representatives, including Japanese encephalitis, West Nile, yellow fever, Dengue (types 1–4), and Zika viruses were extracted and used in the RT-RPA assay.
To assess the performance of this RT-RPA assay in the clinical setting, three serum samples and 17 cerebral spinal fluid (CSF) samples from TBE patients, as well as 15 TBEV-negative samples, were analyzed using the developed RT-RPA assay and the traditional rRT-PCR assay. RNAs were extracted and then subjected to RT-RPA and RT-PCR assays. Both RT-RPA and RT-PCR results were negative for the 15 TBEV-negative samples.
Among the three serum samples from patients with TBEV infection, two samples were TBEV-positive by both RT-RPA and RT-PCR, whereas one sample was negative according to RPA and positive according to RT-PCR. The crossing point (Cp) value of this serum by RT-PCR was 37.16.
For the 17 CSF samples, 14 CSF samples were TBEV-positive by both RT-RPA and RT-PCR; however, three samples yielded inconsistent results with weakly positive results by RT-PCR and negative results by RT-RPA. The Cp values of the three samples were higher than 35 (Table 1).

Table 1. Analysis of clinical samples by RT-RPA and rRT-PCR.
To confirm that TBEV could be detected in the three weakly positive samples by RT-RPA, we repeated the experiment with double the volume of RNA template (10 μL). The results revealed that all three samples were now clearly positive by RT-RPA, suggesting that RT-RPA was less sensitive than RT-PCR. Thus, RT-RPA could probably achieve results similar to those of RT-PCR for samples with Cp values under 35, although the sensitivity might be improved by adding more template RNA.
In summary, we established an RT-RPA assay for TBEV. The LOD of RT-RPA was determined to be 10 copies and 0.1 PFU/reaction. Although the sensitivity of RT-RPA for TBEV was lower than that of rRT-PCR, the assay was easy to perform, and amplification was much more rapid than that in rRT-PCR (RT-RPA, 15 min; rRT-PCR, 2 h); thus, our RT-RPA assay may be useful for detection of TBEV infection under field conditions.
HTML
-
This work was supported by the National Major Science Program Foundation (Nos. 2018ZX10711001-003, 2018ZX10302401-008 and 2017ZX10305501-010), the Major Special Program Foundation (No. AWS15J006), and the National China Science Foundation (No. 81501789).
-
The authors declare that they have no conflict of interest.
-
This study was approved by the ethics committee of Beijing Institute of Microbiology and Epidemiology. Informed consent was obtained from all of the patients whose sera or CSF samples were used in this study.
Conflict of interest
Animal and Human Rights Statement
-
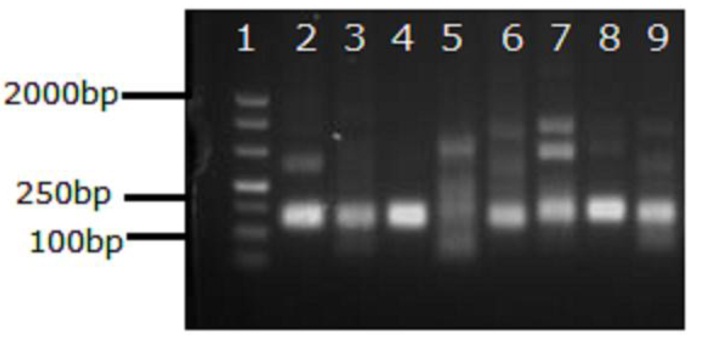
Figure S1. Results of the basic RPA assay performed using primer sets designed for TBEV detection. Lanes: 1, marker DL500bp; 2–9, assay performed using primer sets designed for TBEV detection (Sets 1–8, respectively). A Basic RPA Kit was used to amplify 1000 copies of TBEV VLP RNA. Different primer sets were added to the basic RPA assays. After incubation at 42℃ for 20min, the basic RPA products were visualized by DNA gel electrophoresis. Sets 1–8: set 1, TBEV-F1-RPA and TBEV-R1-RPA; set 2, TBEV-F2-RPA and TBEV-R1-RPA; set 3, TBEV-F3-RPA and TBEV-R1-RPA; set 4, TBEV-F4-RPA and TBEV-R1-RPA; set 5, TBEV-F1-RPA and TBEV-R2-RPA; set 6, TBEV-F2-RPA and TBEV-R2-RPA; set 7, TBEV-F3-RPA and TBEV-R2-RPA; set 8, TBEV-F4-RPA and TBEV-R2-RPA.
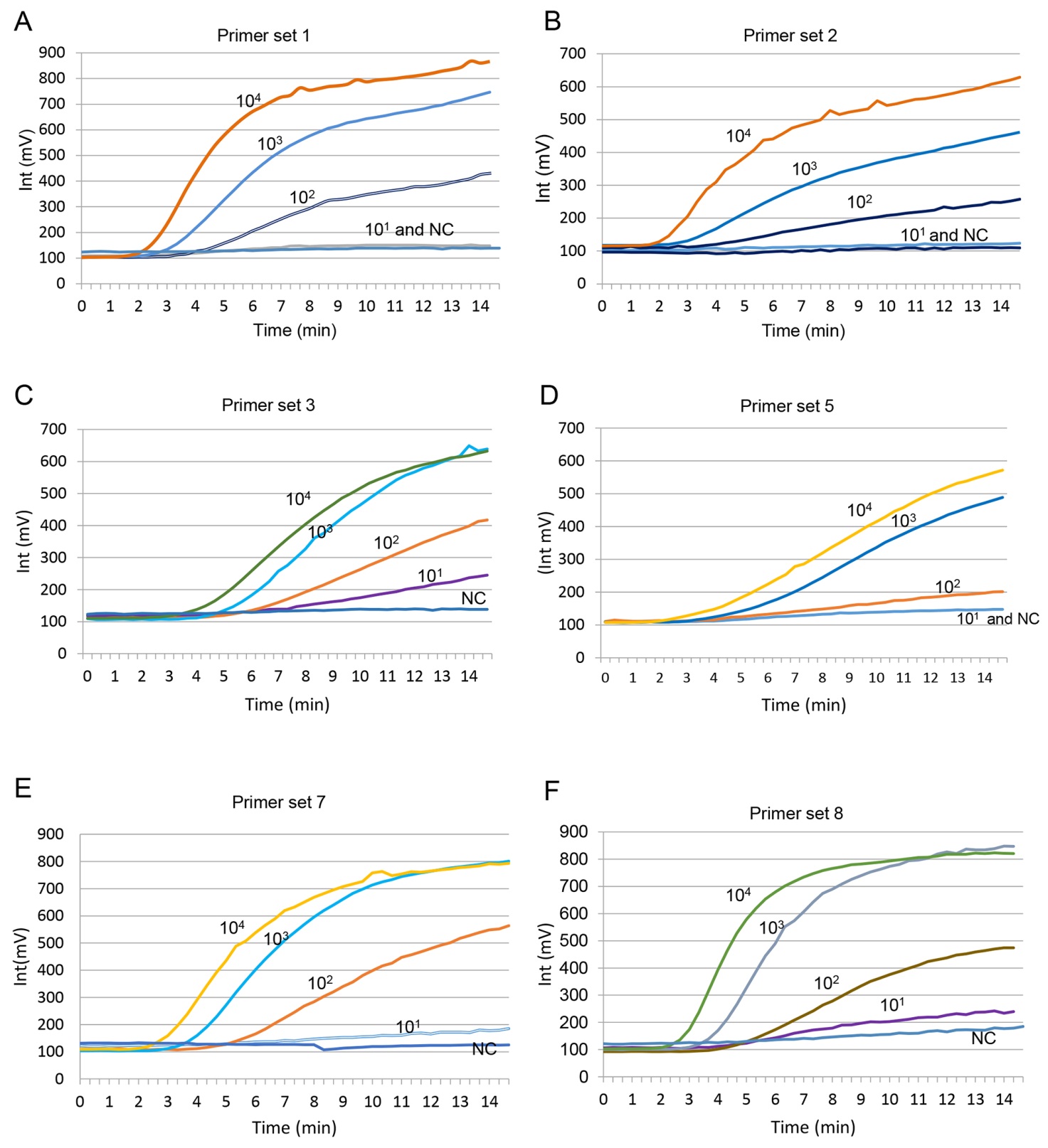
Figure S2. Primer sets and probe screened by RT-RPA. (A) Amplification curve by primer set 1 (TBEV-F1-RPA and TBEV-R1-RPA). (B) Amplification curve by primer set 2 (TBEV-F2-RPA and TBEV-R1-RPA). (C) Amplification curve by primer set 3 (TBEV-F3-RPA and TBEV-R1-RPA). (D) Amplification curve by primer set 5 (TBEV-F1-RPA and TBEV-R2-RPA). (E) Amplification curve by primer set 7 (TBEV-F3-RPA and TBEV-R2-RPA). (F) Amplification curve by primer set 8 (TBEV-F4-RPA and TBEV-R2-RPA).The RNA template from TBEV VLP was 10-fold serially diluted (100 copies/μL to 105 copies/μL) and applied to RT-RPA assay, at 5 μL/reaction; each reaction contained a different primer set and TBEV-RPA-probe and the enzyme complex pellet. After incubating at 39 ℃ for 20 min in the ESE Quant TS, the fluorescence intensity was documented every 20 s, and amplification curves were generated by Tube canner studio software.

Table S1. Sequences of primers and probe for the RPA assay
















 DownLoad:
DownLoad: