HTML
-
Bombyx mori sericulture for silk production is the best developed in China, India, Japan, and Southeast Asia. Grasserie disease, caused by Bombyx mori nucleopolyhedrovirus (BmNPV), is the most common and serious disease in silkworms, accounting for 70%–80% of total losses (Jiang and Xia 2014). BmNPV belongs to the genus Alphabaculovirus in the family Baculoviridae (Harrison et al. 2018). It has a large circular, supercoiled, double-stranded DNA genome of 128, 413 nucleotides containing 141 open reading frames (ORFs) packaged into rod-shaped virions (Gomi et al. 1999; Mikhailov 2003). The main measure to prevent and control BmNPV infection is breeding resistant silkworm varieties via genetic and transgenic breeding (Ponnuvel et al. 2003). However, overexpression of antiviral genes (genes encoding Bmlispase-1, serine protease-2, Sprouty, BmAtlastin-n, and NADH-oxidoreductase-like protein) or RNA interference (RNAi) of BmNPV key genes (e.g., immediate early-1 (ie-1), late expression factor 1 (lef-1), lef-2, lef-3, lef-11, oral infection factor p74, and gp64) by transgene technologies only partially inhibit viral replication and do not completely eliminate the viral genome (Isobe et al. 2004; Jiang and Xia 2014; Ponnuvel et al. 2003; Yao et al. 2008; Zhang et al. 2014a, b). Therefore, these technologies do not meet the needs of the sericulture industry.
A new strategy based on clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 nuclease (CRISPR/Cas9) genome editing has been widely used to eradicate pathogenic viruses, including human immunodeficiency virus 1 (HIV-1), human papillomavirus, hepatitis B virus (HBV), Epstein-Barr virus, plant viruses, and flaviviruses, in infected cells or animal models (Yin et al. 2015; Pellagatti et al. 2015). Over the past 4 years, CRISPR/Cas9 technology has been extensively utilized in basic science and transgenic breeding to treat genetic and infectious diseases in silkworms (Chen et al. 2017; Dong et al. 2018a, 2019). Our laboratory has employed CRISPR/Cas9 technology to develop a BmNPV-specific geneediting system to excise the ie-1 gene from the BmNPV genome in infected B. mori worms (Dong et al. 2016).
Since 2017, targeted viral genes, including ie-0, ie-1, ie-2, and me53, have been edited mainly through singlegene editing, double-gene editing, and large fragment deletion strategies in transgenic breeding (Dong et al. 2018a, 2019). We previously established a virus-inducible CRISPR/Cas9 system with reduced off-target mutagenesis and host toxicity compared to those of conventional CRISPR/Cas9 systems (Dong et al. 2018b). Our system allows not only direct editing of key BmNPV genes, but also editing of host factors upon which the virus depends to inhibit BmNPV multiplication and replication. The application of this technology has yielded remarkable antiviral effects; however, this technology is still inefficient and shows partial selectivity of viral gene editing for BmNPV genome removal (Chen et al. 2017; Dong et al. 2019). Thus, there is an urgent need to screen and identify BmNPV genes for potential use in a multigene-editing system to improve antiviral efficacy via multigene editing of the BmNPV genome.
To achieve highly specific BmNPV genome editing, we created an efficient multiplex CRISPR/Cas9 gene-editing technology to disrupt the BmNPV genome in infected silkworm cells. We first constructed a one-vector CRISPR/Cas9 gene-editing plasmid, pSL1180-Cas9-U6-single guide (sg)RNA, using BmNPV lef-11 as a target gene. This plasmid can be used to quickly insert target genes and detect antiviral activity. In addition, we screened ie-1 and gp64 genes, which are important for viral DNA replication and systemic infection. More importantly, to completely eliminate the BmNPV genome from infected silkworm cells, we constructed an anti-BmNPV therapeutic multiplex (PSL1180-Cas9-sgIE1-sgLEF11-sgGP64) CRISPR/Cas9 system. We identified a series of highly active sgRNA candidates that would be optimal for therapeutic application. Using the multiplex CRISPR/Cas9 genome-editing system, we were able to inhibit BmNPV replication.
-
The B. mori ovary cell line BmN-SWU1 was maintained in TC-100 medium supplemented with 10% (v/v) fetal bovine serum (FBS) (Thermo Fisher Scientific, MA, USA) and 10% (v/v) penicillin/streptomycin at 27 ℃ (Pan et al. 2010). An enhanced green fluorescent protein marker gene (EGFP) driven by the BmNPV 39K promoter (v39Kprm-EGFP) was inserted into the polyhedrin locus of wild-type virus by Tn7-mediated transposition using the Bac-to-Bac system (Thermo Fisher Scientific, MA, USA) (Dong et al. 2014; Zhang et al. 2014a). Recombinant BmNPV particles were generated in BmN-SWU1 cells and viral titers were determined using the 50% tissue culture infective dose assay (TCID50) (Dong et al. 2014).
-
SgRNA primer pairs targeting the amino-terminal regions of ORFs of key BmNPV genes (ie-0, ie-1, ie-2, lef-1, lef-3, lef-11, gp64, vp39, poly, p143, p35, and DNApol) were designed using the CRISPR website (http://crispr.dbcls.jp/) (Naito et al. 2015). To efficiently identify target genes, we scanning for GN19NGG sequences in the BmNPV genome, and identified anti-BmNPV sgRNA candidates that met target sequence requirements and had their own PAM recognition domains. All sgRNAs scaffold target sequences were inserted upstream and downstream of the BbsI site. In addition, we synthesized multiple sgRNAs at the Beijing Genomics Institute (BGI, Beijing, China). The primer sequences are listed in Supplementary Table S1.
-
Cloning of the Cas9 gene and preparation of the expression cassette for sgRNA were conducted as described previously (Dong et al. 2016). Briefly, to generate a pSL1180- IE1-Cas9-Ser plasmid that encodes the target gene along with Cas9 and a Flag epitope, the Cas9 sequence was PCRamplified from previously established vector construct and inserted behind the BmNPV IE-1 promoter in the B. mori sericin polyA-based pSL1180 vector. The U6 promoter and sgRNA were ligated into the vector following single digestion of pSL1180-IE1-Cas9-Ser-PA with EcoR I. The resulting Cas9 and sgRNA expression vector was named pSL1180-IE1-Cas9-Ser-U6-sgRNA. We linked the sgIE1- sgLEF11-sgGP64 expression cassette into the pSL1180- IE1-Cas9-Ser vector and used restriction enzymes to verify correct cloning. The final construct was named PSL1180- Cas9-sgIE1-sgLEF11-sgGP64 (sgMultiple). All cloneswere verified by sequencing. Primers used are listed in Supplementary Table S2.
-
BmN-SWU1 cells (1×105) were seeded on cover slips (Thermo Fisher Scientific, MA, USA) in 24-well plates and cultured at 27 ℃ for 48 h such that they reached 70% confluence. After cell stabilization, the plasmid was transfected into the cells using the X-tremeGENE HP DNA Transfection Reagent (Roche, Basel, Switzerland). At 48 h post-transfection (h p.t.), the cells were infected with BmNPV at a multiplicity of infection (MOI) of 10. At 72 h post infection (h p.i), the BmN-SWU1 cells were subjected to immunofluorescence. Briefly, at 72 h p.i. with BmNPV after transfection of the indicated plasmid, cells were collected and washed three times with phosphate-buffered saline (PBS). The cells were then fixed in cold 4% paraformaldehyde in PBS for 15 min and permeabilized with 1% Triton X-100 for 15 min. After blocking in 3% BSA and 10% sheep serum in PBS at 37 ℃ for 1 h, the cells were incubated with a monoclonal anti-Flag antibody diluted 1:200 in PBS at 37 ℃ for 1 h. Finally, after six washes for 5 min each in PBS, the cells were incubated with Alexa 555-conjugated goat anti-mouse IgG (1:500; Thermo Fisher Scientific, MA, USA) and DAPI (1:500; Thermo Fisher Scientific, MA, USA) for 1 h. They were then washed six times with PBS and observed and imaged under a confocal microscope (FV3000, Olympus, Tokyo, Japan).
-
Total DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Wisconsin, USA). The BmNPV genomic region surrounding the sgRNA target site of each gene was amplified by PCR for detection of cleavage of the lef-11, ie-1, and gp64 genes. PCR products were purified and ligated into the pEASY-T5 vector (TransGen Biotech, Beijing, China) and sequenced with M13 primers (BGI, Beijing, China).
-
Transfected cells were lysed in western and immunoprecipitation buffer (pH 7.5) containing 20 mmol/L Tris–HCl, 150 mmol/L NaCl, and 1% Triton X-100 (Beyotime, Shanghai, China). The lysates were mixed with loading buffer (5×) and boiled for 5 min. The protein samples were then separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After blocking with 5% skim milk, the membranes were incubated with an antirabbit VP39 polyclonal antibody (1:2000) and an antirabbit tubulin polyclonal antibody (1:5000; Sigma, CA, USA) in 5% milk in Tris-buffered saline containing Tween-20 at 27 ℃ for 1 h. After washing, the blot was incubated with a horseradish peroxidase-labeled goat antirabbit IgG (1:20, 000; Beyotime, Shanghai, China) at 37 ℃ for 1 h. Clarity Western ECL Substrate (Bio-Rad, CA, USA) was added and the blot was exposed to X-ray film for detection.
-
BmNPV genomic DNA was subjected to qPCR using SYBR Premix Ex Taq (TaKaRa, Dalian, China) following a previously described protocol and using reported gp41 specific primers (Dong et al. 2017). For quantification, a DNA standard curve was constructed based on Ct values to calculate gp41 gene copy number as previously described (Dong et al. 2014).
-
At 48 h post transfection, cells were infected with BmNPV at an MOI of 10. The cells were collected in PBS at the indicated time points after infection. The percentage of EGFP-positive cells was determined using a NovoCyte flow cytometer (ACEA Biosciences, MA, USA). The data were analyzed using the Novo Express software.
-
All data are presented as the mean ± standard deviation. Unpaired, one-tailed Student's t-tests were used to compute differences between two independent groups in GraphPad Prism 6 (GraphPad Software, Inc., CA, USA). P < 0.01 was considered statistically significant.
Cells and Viruses
sgRNA Design
Plasmid Construction
Immunofluorescence
Sequence Analysis
Western Blotting
Quantitative PCR (qPCR) for DNA Replication Assay
Flow Cytometry
Statistical Analysis
-
BmNPV contains 141 genes, most of which have not been studied previously (Ono et al. 2012). To evaluate the effect of knockout of many of these genes on BmNPV replication, we constructed a one-vector CRISPR/Cas9 system for rapid detection of BmNPV multiplication inhibition. We linked Cas9 to a U6-sgRNA expression cassette into the pSL1180 vector (Fig. 1A). We previously demonstrated that the BmNPV lef-11 gene is necessary for virus replication (Dong et al. 2015). Here, we first evaluated gene editing and antiviral efficacy of the system by knocking out lef-11 using three different target sites in lef-11. At 72 h post infection, the genome copy number of BmNPV was measured by qPCR. The cells treated with sgLEF11-1 (Targeting the 30 UTR region of LEF-11), sgLEF11-2 (Targeting the ORF region of LEF-11), and sgLEF11-3 (Targeting the 50 UTR region of LEF-11) had lower levels of BmNPV DNA than cells transfected with sgMock. DNA levels in cells transfected with sgLEF11-2 were 10-fold lower than those in sgMock control cells (Fig. 1B).
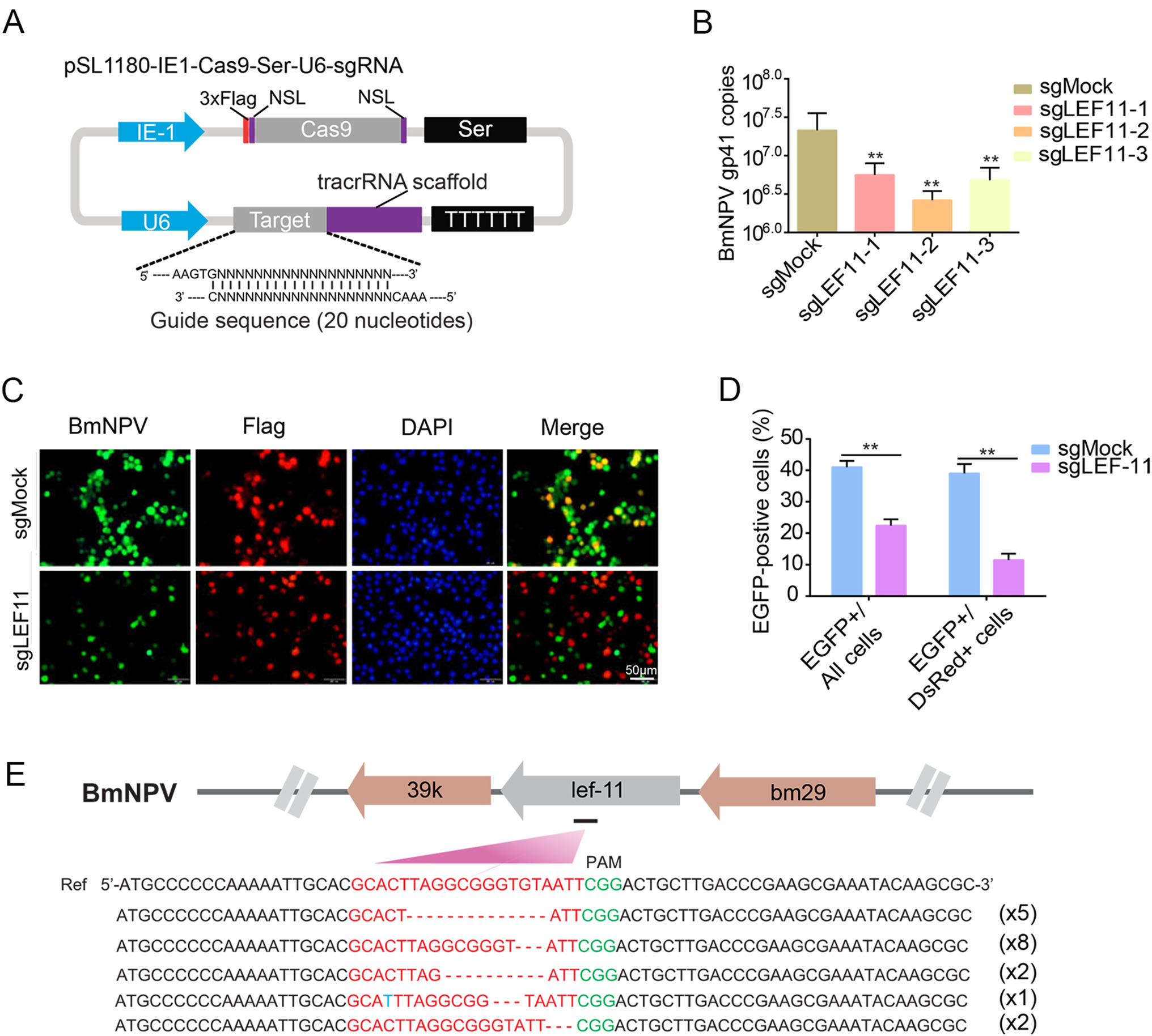
Figure 1. A one-vector CRISPR/Cas9 system for BmNPV genome excision. A Schematic representation of the one-vector CRISPR/Cas9 system constructed in this study. A single 20-bp sgRNA sequence was inserted downstream of the U6 promoter into pSL1180 using the BbsI restriction site. B BmNPV gp41 copy numbers in cells treated with sgLEF11-1, sgLEF11-2, and sgLEF11-3 as determined by qPCR. **P < 0.01. C Fluorescence microscopic images of BmN-SWU1 cells transfected with the one-vector CRISPR/Cas9 system followed by infection with BmNPV for 72 h. Cas9 protein was labeled with Alexa 555-conjugated goat anti-mouse IgG (red) and green fluorescence represents BmNPV-infected cells. Scale bar = 50 μm (D) BmNPV infection rates were determined as EGFP+ cells in all cells and Cas9-expressing (DsRed+) cells counted in five fields. Three independent experiments were carried out in triplicate. **P < 0.01. E DNA sequence alignment of the CRISPR/Cas9 lef-11 target sites. The wild-type reference sequence is shown at the top. The target sequences of lef-11 are indicated in red. Deletions are indicated by dashes. The numbers in parentheses represent the number of occurrences of the different lef-11 gene deletions.
Forty-eight hours after transfection with sgLEF-11 (sgLEF11-2), BmN-SWU1 cells were infected with v39Kprm-EGFP for 72 h. Inhibition of virus replication was evaluated by immunofluorescence. The CRISPR/Cas9 system expressed the Cas9 protein with a Flag tag, and red fluorescence could be observed after the cells were stained using a monoclonal anti-Flag antibody and an Alexa 555-conjugated goat anti-mouse IgG antibody. Green fluorescence was observed from the recombinant virus harboring v39Kprm-EGFP. Most cells transfected with sgLEF- 11 did not have detectable EGFP levels, whereas EGFP signals were clearly detected in transfected sgMock cells (Fig. 1C). Quantitative analysis revealed that the number of EGFP+ cells decreased significantly compared with the sgMock from 41% to 20% upon transfection with sgLEF- 11. There was a similar marked reduction in the number of EGFP+/Flag+ cells, from 40% to 12%, in cells transfected with sgLEF-11 (Fig. 1D). Sequence analysis of the lef-11 region showed that EGFP expression was disrupted by various mutations, mostly 3–12-bp deletions near the putative target site (Fig. 1E). These results indicated that the one-vector CRISPR/Cas9 system can be used to rapidly identify key genes that inhibit BmNPV replication based on genome copy number and fluorescence of a reporter molecule.
-
The genes ie-1, lef-3, p143, lef-1, and DNApol are essential for BmNPV DNA replication, although the exact functions of most DNA replication-related genes in BmNPV are unknown (Vanarsdall et al. 2007). Based on the principle that early expressed genes in BmNPV have a greater influence on viral DNA replication, we selected ie-1, gp64, lef-3, lef-11, ie-2, p35, p143, lef-1, and DNApol genes as candidate target genes to use in our one-vector CRISPR/Cas9 system. To evaluate the efficacy of the selected sgRNAs in targeting BmNPV, we transfected the onevector CRISPR/Cas9 system into BmN-SWU1 cells. At 72 h post virus infection, the cells were analyzed by immunofluorescence. As shown in Fig. 2A, sgIE-1, sgGP64, sgLEF-3, sgDNApol, and sgLEF-11 inhibited EGFP expression more effectively than sgLEF-1, sgP143, sgIE-2, sgP35, and sgMock. Quantitative analysis showed that the EGFP+ cells among totals cells transfected with sgIE-1, sgGP64, sgLEF-3, sgDNApol, or sgLEF-11 were only 20%–30%, versus 50% for sgMock-transfected cells (Fig. 2B). Analysis of BmNPV DNA replication revealed that sgIE-1, sgGP64, and sgLEF-11 were the most effective at inhibiting viral DNA replication among all target genes. The copy numbers of BmNPV gp41 in cells transfected with sgIE-1, sgGP64, or sgLEF-11 were 10-fold lower than those in cells transfected with sgMock (Fig. 2C). Flow cytometric analysis revealed that the numbers of EGFP+ cells among total cells transfected with sgIE-1, sgGP64, or sgLEF-11 were lower than that in sgMock cells (Fig. 2D). The fraction of EGFP+ cells significantly decreased from 65.36% to 45.97%, 47.35%, and 46.07% after transfection with sgIE-1, sgGP64, or sgLEF-11, respectively (Fig. 2D).
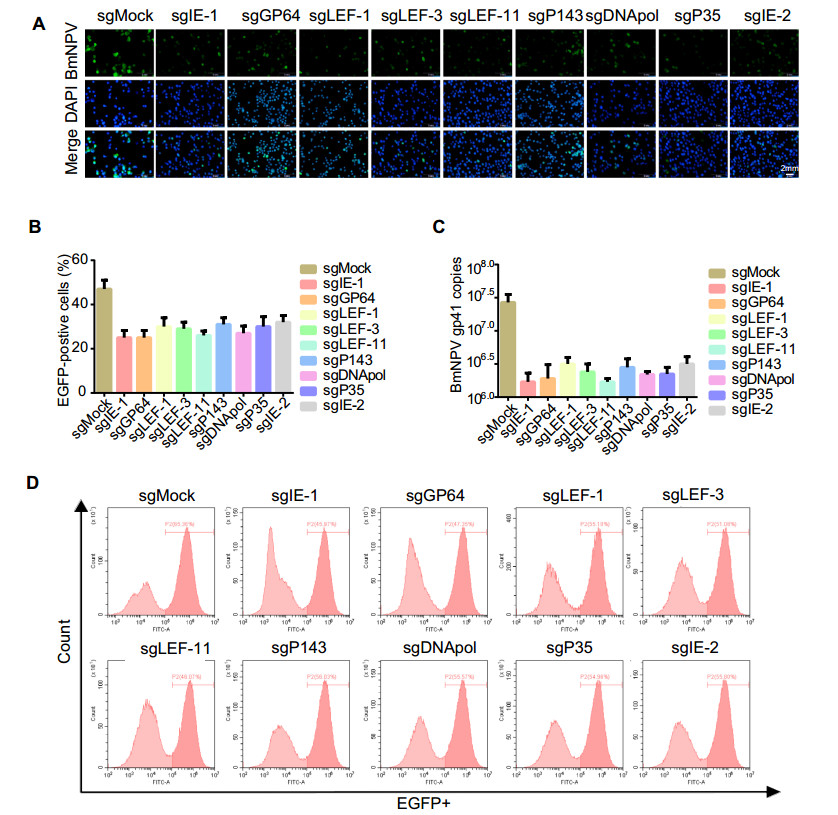
Figure 2. Screening of key genes involved in BmNPV DNA replication. A Antiviral efficiency of knockout of genes involved in BmNPV DNA replication. BmN-SWU1 cells were transfected with the indicated sgRNAs (sgMock, sgIE-1, sgLEF-1, sgLEF-3, sgLEF-11, sgP143, sgDNApol, sgP35, and sgIE-2) and infected with BmNPV at an MOI of 10. Light and fluorescence microscopic images were acquired after co-culturing the cells with BmNPV for 72 h. B EGFPpositive cell fractions among BmN-SWU1 cells transfected with Cas9 and the indicated sgRNA. C BmN-SWU1 cells were transfected with sgMock, sgIE-1, sgLEF-1, sgLEF-3, sgLEF-11, sgP143, sgDNApol, sgP35, or sgIE-2 and infected with BmNPV at an MOI of 10. At 72 h p.i., total DNA was extracted from each Cas9-transfected group and viral gp41 copies were quantified by qPCR. D Flow cytometry analysis was used to quantify BmNPV replication at different time points in each set of cells transfected with sgRNA and infected with BmNPV.
-
Next, we tested whether a combination of two sgRNAs or multiple sgRNAs could more effectively inhibit viral replication. Thus, one CRISPR/Cas9 genome editing system contains multiple sgRNAs that the Cas9 protein edits simultaneously (Schiwon et al. 2018). First, we transduced cells with single and double sgRNAs via our one-vector system to disrupt BmNPV infection. Double sgRNAs inhibited viral infection more effectively than single sgRNAs (Supplementary Figure S1). Next, we selected ie-1, gp64, and lef-11 as the target genes for multigene editing via a systematic screening combined with the baculovirus infection process. The sgRNA and Cas9 expression cassettes were inserted into a one-vector system named sgMultiple (Fig. 3A). Sequence analysis of the ie-1, gp64, and lef-11 regions showed that BmNPV replication was disrupted due to various mutations and deletions near the putative target sites (Fig. 3A).

Figure 3. CRISPR/Cas9-mediated multigene editing impairs BmNPV replication. A Schematic representation of the CRISPR/Cas9 multigene-editing vector. DNA sequence alignment of the CRISPR/Cas9 target sites in the ie-1, gp64, and lef-11 regions. The wild-type sequence of each region is shown on top, the target sequence is indicated in blue, and deletions are indicated by dashes. B BmNSWU1 cells were transfected with multigene-editing vectors, infected with BmNPV, and examined by fluorescence microscopy at 72 h p.i. Cas9 protein was labeled with Alexa Fluor 555 (red), green fluorescence represents BmNPV-infected cells, and nuclei were stained with DAPI (blue). SgMock represents a non-target sequence, and sgMultiple represents the four target sequences of this multiplex gene editing system. Scale bar = 50 μm. C BmNPV infection rates determined based on the number of EGFP-positive cells in multiplex gene target cells and in non-target cells counted in five different fluorescent fields. All experiments were repeated three times. **P < 0.01. D BmNPV multiplication was measured in sgMockand sgMultiple-transfected cells infected with BmNPV at different time points by flow-cytometric analysis. (E) EGFP+ cells among BmN-SWU1 cells transfected with sgMock or sgMultiple and infected with BmNPV. **P < 0.01.
Fluorescence microscopy revealed that cells stably expressing sgMultiple (red) significantly inhibited virus replication compared with sgMock expressing cell with BmNPV (green) 72 h p.i. (Fig. 3B). Quantitative analysis revealed that the fractions of EGFP+ cells decreased significantly from 40%–45% to 18%–23% after transfection with sgMultiple. In addition, there was a marked reduction in the fraction of EGFP+/Flag+ cells from 40% to 3% in sgMultiple-transfected cells (Fig. 3C). Flow cytometry was used to further analyze the efficiency of BmNPV proliferation inhibition by sgMultiple. sgMultiple significantly inhibited BmNPV multiplication compared with sgMock at different time points post infection (Fig. 3D). Quantitative analysis revealed that the fraction of EGFP+ cells decreased significantly from 57.89% to 9.26% after transfection with sgMultiple (Fig. 3E). These results indicated that the simultaneous use of multiple sgRNAs can effectively increase the efficacy of viral replication inhibition.
-
We also analyzed viral DNA replication in sgMultipletransfected cells. At 48 h p.i., the copy number of BmNPV was 75% lower in sgMultiple-transfected cells than in sgMock-transfected cells (Fig. 4A). To confirm that the multigene-editing system could effectively inhibit virus proliferation, we analyzed changes in the budded virus titer by TCID50 assays. After transfection with sgMultiple and sgMock, we measured the viral titer in the cells. The viral titer in sgMultiple-transfected cells was 100-fold lower than that in sgMock-transfected cells (Fig. 4B). Finally, we analyzed the effect of multigene editing on the expression of the BmNPV VP39 protein. VP39 expression was significantly lower in sgMultiple-transfected cells than in sgMock-transfected cells (Fig. 4C). These results suggested that the CRISPR/Cas9-mediated multigene-editing system effectively impaired BmNPV replication and can be applied in antiviral research in transgenic silkworms.
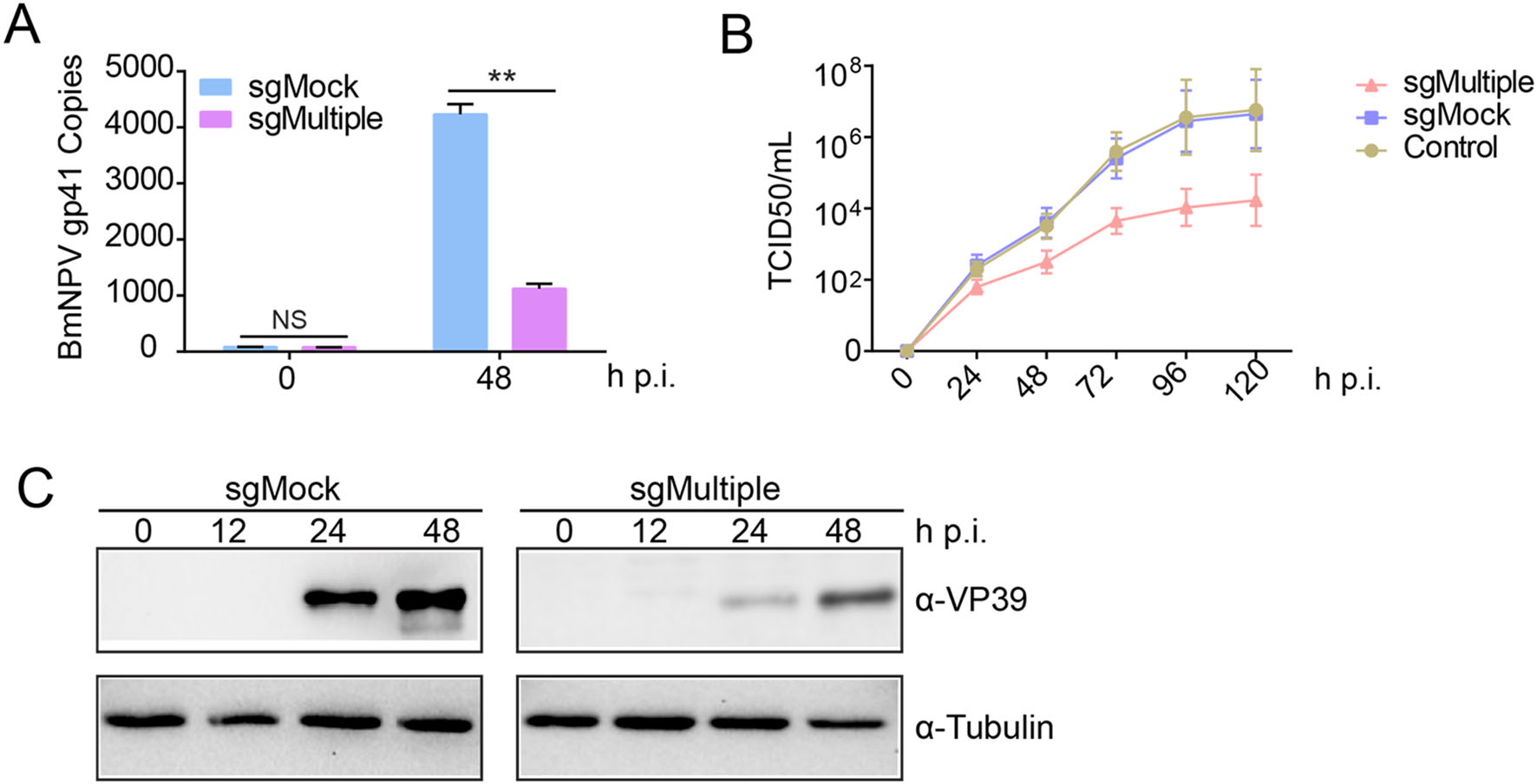
Figure 4. Antiviral efficacy of the CRISPR/Cas9-mediated multigeneediting system A Viral DNA expression in sgMultiple cells infected with BmNPV at an MOI of 10. At 48 h p.i., total DNA was extracted from the transfected cells and viral gp41 copies were quantified by qPCR. Data are the mean from three independent replicates. **P < 0.01. NS, not significant. B Viral growth was determined by TCID50 assays. Fluorescence was measured after 24, 48, 72, 96, and 120 h p.i. C Antiviral activity of sgMultiple was monitored based on the protein levels of the late expression portion VP39 (top) measured by Western blotting. Tubulin (bottom) was used as a loading control.
Establishment of a One-vector CRISPR/Cas9 System for BmNPV Genome Excision
Screening of Key Genes Involved in BmNPV DNA Replication
CRISPR/Cas9-mediated Multiplex Gene Editing Impairs BmNPV Replication
Antiviral Efficacy of the CRISPR/Cas9-mediated Multiplex Gene-editing System
-
CRISPR/Cas9-mediated gene editing has been widely used in insect pest control, sex regulation, silk protein synthesis, and infectious disease research (Ma et al. 2018). Previously, we developed a series of transgenic silkworm lines with high antiviral ability via transgenic gene editing (Dong et al. 2018a, b). Here, we successfully disrupted BmNPV replication by utilizing a newly established multiplex CRISPR/Cas9 system. To the best of our knowledge, disruption of multiple steps of viral infection with the CRISPR/Cas9 gene-editing system has not yet been demonstrated in insect cells.
A single Cas9 protein can load multiple sgRNAs; therefore, the CRISPR/Cas9 system can effectively knock out multiple target genes (Jang et al. 2018; Zhang et al. 2016). This has allowed the identification of genetic traits of multiple genes and knockout of gene families (Jang et al. 2018; Schiml and Puchta 2016). Multigene-editing tools have been widely used to construct multigenedelivery systems to deliver multiple sgRNAs for the treatment of HIV, retrovirus, lentivirus, and HBV infections (Hu et al. 2017; Sakuma et al. 2016; Schiwon et al. 2018; Yin et al. 2018). Here, we constructed a one-vector CRISPR/Cas9 system that allows the screening and identification of key genes involved in BmNPV systemic infection as well as the verification of its antiviral efficacy (Fig. 4). This technology may be valuable in protecting silkworms by eliminating viral infection at multiple stages.
In 2004, the BmNPV lef-1 gene was targeted by RNAi to inhibit viral replication (Isobe et al. 2004). Since then, multiple other genes involved in BmNPV systemic infection have been modulated, including microRNA, dsRNA, and shRNA (Zhang et al. 2014a, b; Zhou et al. 2014); however, the target genes used in these methods were different from those considered in our approach. Therefore, systematic comparison of the antiviral efficacy of the genes was not possible. Here, 11 genes with different functions in different stages of BmNPV infection were targeted using the one-vector CRISPR/Cas9 system (Figs. 2 and 3). The antiviral ability of sgRNAs targeting these genes was systematically compared by assessing DNA replication and viral fluorescence. The screening yielded various reliable target genes for future development of transgenic antiviral silkworms. It also highlights future research directions to elucidate BmNPV gene functions. In addition to BmNPV, this method can be used to screen and edit target genes in other viruses, such as Bombyx mori cypovirus, Bombyx mori densonucleosis virus, and Bombyx mori flacherie virus (Cao et al. 2012; Li et al. 2001).
Other approaches can be used to generate transgenic silkworms that can resist bacterial diseases, fungal diseases, and Nosema bombycis, depending on the different characteristics of silkworm diseases. Recently, the CRISPR family gene-editing systems include CRISPR/CPF1 and CRISPR/Cas13, which can edit DNA, RNA, or both DNA and RNA sequences (Mahas and Mahfouz 2018; Tsukamoto et al. 2018). CRISPR/CPF1 and CRISPR/Cas13 systems have smaller nuclease proteins and have more RNA target sequences and lower off-target efficiency. Then these can be systematically applied to research on silkworm disease resistance breeding and the results may benefit the sericulture industry (Konermann et al. 2018).
In conclusion, the multiplex CRISPR/Cas9 system constructed in this study has high gene editing efficiency and antiviral ability and may present a strategy for antiBmNPV therapy. In future, the application of this multiplex CRISPR/Cas9 genome-editing technology may provide new insights to improve the strategic inhibition of multiple silkworm pathogens simultaneously. CRISPRmediated multigene editing can be used to breed transgenic antiviral lines and in insect pest control.
-
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31872427 and 31572466), China Agriculture Research System (CARS-18), Chongqing Special Postdoctoral Science Foundation (XmT2018020), and China Postdoctoral Science Foundation (2018M633309).
-
ZD and ZH conceived and designed the experiments. LH, QQ, TT and XZ curated the Data. ZD, PC and QQ analyzed the date; ZD, CL and MP contributed to the writing of manuscript. All authors reviewed and approved the final manuscript for submission.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: