HTML
-
Nucleos(t)ide analogs (NAs) can efficiently inhibit hepatitis B virus (HBV) replication but may select drug-resistant HBV isolates with mutations (MTs) within the open reading frame of polymerase (Zoulim and Locarnini 2009; Lok et al. 2016). Due to the overlapping nature of the open reading frames of HBV genomes, resistance MTs in polymerase may affect hepatitis B surface proteins, leading to alterations in virion secretion ability, infectivity, vaccine efficacy, liver disease pathogenesis and transmission throughout the population. For example, the rtA181T nucleotide substitution in the polymerase open reading frame may result in the sW172* (stop codon) substitution in the S domain, which has a dominant-negative effect of secretion and increases oncogenic potential (Yeh 2010). Compared to the rtA181T surface missense MT (rtA181T/sW172S), introduction of the rtA181T surface nonsense MT (rtA181T/sW172*) caused decreased viral replication and altered drug resistance, suggesting that overlapping changes in the surface gene may potentially affect the impact of HBV polymerase MT on replication and drug resistance (Ahn et al. 2014; Zhao et al. 2018). Therefore, more attention should be given to NA resistance MTs that result in a stop codon MT in the envelope gene.
NA resistance MTs are usually detected as a mixed population with wild-type (WT) HBV (Warner and Locarnini 2008; Zoulim and Locarnini 2009; Colledge et al. 2017). However, there is scant information on how the coexistence of NA resistance MTs and WT HBV affects HBV replication. Considering that NA resistance MTs with a corresponding stop codon MT in the envelope gene result in secretion deficiency, this scenario can be readily applied to investigate the complementary relationship between NA resistance MTs and WT. We hypothesized that WT HBV and drug-resistant HBV would complement with each other to maintain viral replication and rescue virion production under NA treatment.
In this study, three most frequently detected NA resistance MTs (rtA181T/sW172*, rtV191I/sW182* and rtM204I/sW196*) that result in a stop codon MT in the envelope gene (Colledge et al. 2017), were transiently transfected into Huh7 cells alone or together with their corresponding WT in different ratios in the absence or presence of NAs. Their impact on viral replication and secretion were characterized by using Western blotting and Southern blot analysis. These results will help to reveal how the coexistence of NA resistance MTs and WT HBV affects HBV replication.
-
The complete RT gene with rtA181T/sW172*, rtV191I/sW182*, or rtM204I/sW196* MT or their corresponding WT RT gene, WT(sW172*), WT(sW182*) or WT(sW196*), were amplified from clinical isolates and cloned into the pTriEx-mod-1.1 vector after sequencing, as described previously (Liu et al. 2011, 2014). Thus, replication-competent clones pHBV-MTs including pHBV-sW172*, pHBVsW182*, and pHBV-sW196* and their corresponding plasmids containing WT HBV, pHBV-WT(sW172*), pHBV-WT(sW182*) and pHBV-WT(sW196*) were generated.
-
Human hepatoma Huh7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin. For cell transfection, Huh7 cells were seeded at approximately 30% confluence and transfected with Lipofectamine 2000 according to the manufacturer's instructions as described previously (Wu et al. 2012; Qin et al. 2013). Huh7 cells were harvested at 72 h posttransfection. Western blotting was performed as described previously (Li et al. 2015). The following primary antibodies were used: anti-HBs (Abcam) and anti-β-actin (Santa Cruz). Horseradish peroxidase (HRP)-conjugated antibodies (Jackson) were used as secondary antibodies. HBsAg in cell culture supernatants were detected by ELISA, as described previously (Zhao et al. 2017).
-
Encapsidated HBV replication intermediates were extracted from Huh7 cells at 72 h post-transfection and subjected to Southern blot analysis, as described previously (Meng et al. 2008; Zhang et al. 2011; Qin et al. 2013; Cao et al. 2014), which is a well-established method for HBV research. In brief, transfected cells were first lysed in cell lysis buffer. Nuclei were removed by centrifugation. The supernatant was treated with DNase I to digested free DNA (all transfected plasmids have to be digested at this step). Then HBV capsids were digested with proteinase K. Finally HBV capsid-associated DNA was purified by phenol/chloroform extraction. After that, the isolated HBV DNA was subjected to agarose gel electrophoresis, followed by denaturation and Southern blotting. HBV DNA was detected by hybridization with a 32P-labeled full length HBV DNA probe. Hybridization signals were visualized and analyzed by a Phospho-Imager. In addition, hybridization signals were quantified with ImageJ software (National Institutes of Health).
-
For the IF assay, Huh7 cells were seeded at approximately 30% confluence in 6-well plates and transfected with 1.5 μg plasmid. At 48 h post-transfection, Huh7 cells were harvested and plated on glass cover slips for 24 h. Then indirect IF staining of transfected cells was performed as previously described (Wu et al. 2010, 2012; Xu et al. 2012). Mouse monoclonal anti-HBs antibody S1 recognizing HBsAg 'a' determinant (amino acid (aa) 124–147) (kindly provided by Prof. Bing Yan) (Wu et al. 2012) was used as the primary antibody and Alexa Fluor 488-conjugated (Life Technologies, Carlsbad, CA) as the secondary antibody.
-
HBV virions were immunoprecipitated from the transfection supernatant using a mixture of three monoclonal antiHBs antibodies, anti-BJ11, anti-S1 and anti-S39 (kindly provided by Prof. Bing Yan), as previously described (Yao et al. 2018). To exclude the contamination of input plasmids, immunoprecipitated virions were pre-treated with 100 mg/mL DNase I (Promega) overnight and virionassociated DNA was extracted by using QIAamp DNA Blood Mini Kit (QIAGEN, Germany) according to the manufacturer's instructions and subjected to Southern blot analysis or real-time PCR as previously described (Zhang et al. 2011).
-
Statistical analysis was carried out using GraphPad (GraphPad Software). Differences in multiple comparisons were assessed for significance using Student's t test. A P value < 0.05 was considered significant. Results are presented as Mean ± S.D.
HBV RT Gene Amplification, Sequencing and Construction of Viral Amplicons Containing the 1.1-mer HBV Genome
Cell Transfection, Western Blotting and EnzymeLinked Immunosorbent Assay (ELISA)
Southern Blot Analysis
Immunofluorescence (IF) Staining and Confocal Laser Scanning Microscopy
Immunoprecipitation of HBV Virions and RealTime Polymerase Chain Reaction (PCR) Analysis
Statistical Analysis
-
In addition to leading to an aa substitution in the RT region of Pol, the rtA181T/sW172*, rtV191I/sW182* and rtM204I/sW196* MTs resulted in a truncation of 55 aa, 45 aa and 31 aa, respectively, in all three forms of HBV surface proteins including large (L), middle (M) and small (S) surface protein (Fig. 1A), which correspond to a decrease in molecular weight of approximately 6 kDa, 4 kDa and 2 kDa, respectively. Thus, expression of the three forms of HBV surface proteins was examined. Compared to their corresponding WT sequences, truncated S proteins for both sW172* and sW182* were barely detectable (Fig. 1B, Lane 1 to 2, Lane 3 to 4). Conversely, both glycosylated and unglycosylated forms of truncated S proteins (St), corresponding to bands at approximately 25 kDa and 22 kDa, respectively, could be observed for sW196*, with a significant decrease (Fig. 1B, Lane 6 to 7). The two glycosylated forms of the M protein were Fig. 1 The RT/surface truncation mutation impaired expression and secretion of HBV surface proteins. A Schematic representation of the RT/surface truncation mutation used in this study. The amino acid substitutions in the reverse transcriptase domain of HBV polymerase (Pol) and their corresponding stop codon mutation in the overlapping surface protein (S) are indicated. Regions that are consequently not translated are indicated by white boxes with dotted lines. Huh7 cells were seeded at approximately 30% confluence in 6-well plates and transfected with 1.5 μg pHBV-sW172* (sW172*), pHBV-sW182* (sW182*), or pHBV-sW196* (sW196*) and corresponding plasmids containing WT HBV, pHBV-WT(sW172*) (WT(sW172*)), pHBVWT(sW182*) (WT(sW182*)) or pHBV- WT(sW196*) (WT(sW196*)), respectively. Cells were collected at 72 h posttransfection. B Expression of HBV surface proteins (L, M, S) and truncated S proteins (St) was detected by Western blotting with an anti-HBs antibody. Expression of b-actin was detected as a loading control. C HBsAg levels in the culture medium from the cells were diluted tenfold and detected by ELISA. detectable in lysates of both WT (sW182*)- and WT (sW196*)-transfected cells but were undetectable in lysates of sW182* and sW196*-transfected cells (Fig. 1B Lanes 3 to 4, Lanes 5 to 6, indicated by the "→" symbol). Similar results were obtained for the L protein (Fig. 1B Lanes 3 to 4, Lanes 5 to 6, indicated by the "*" symbol). Consistent with intracellular results (Fig. 1B), the level of HBsAg was barely detected in the culture supernatants of cells carrying the sW172* and sW182* MT (Fig. 1C). Notably, although intracellular truncated S proteins were detected (Fig. 1B, Lane 6), extracellular HBsAg was not be detected for sW196* (Fig. 1C), indicating a secretion deficiency of the truncated HBsAg proteins.
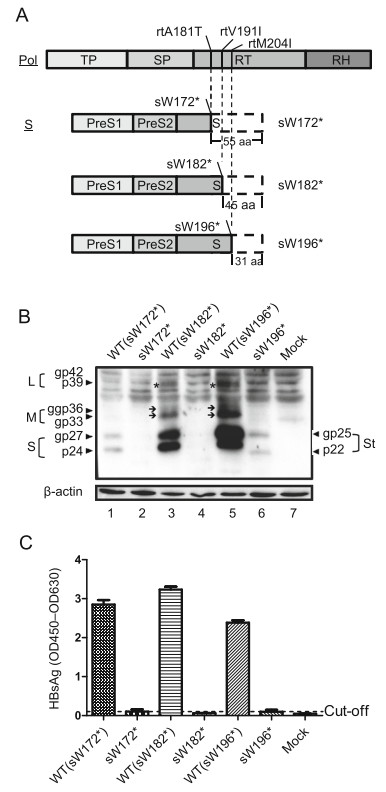
Figure 1. The RT/surface truncation mutation impaired expression and secretion of HBV surface proteins. A Schematic representation of the RT/surface truncation mutation used in this study. The amino acid substitutions in the reverse transcriptase domain of HBV polymerase (Pol) and their corresponding stop codon mutation in the overlapping surface protein (S) are indicated. Regions that are consequently not translated are indicated by white boxes with dotted lines. Huh7 cells were seeded at approximately 30% confluence in 6-well plates and transfected with 1.5 μg pHBV-sW172* (sW172*), pHBV-sW182* (sW182*), or pHBV-sW196* (sW196*) and corresponding plasmids containing WT HBV, pHBV-WT(sW172*) (WT(sW172*)), pHBVWT(sW182*) (WT(sW182*)) or pHBV- WT(sW196*) (WT(sW196*)), respectively. Cells were collected at 72 h posttransfection. B Expression of HBV surface proteins (L, M, S) and truncated S proteins (St) was detected by Western blotting with an anti-HBs antibody. Expression of b-actin was detected as a loading control. C HBsAg levels in the culture medium from the cells were diluted tenfold and detected by ELISA.
Furthermore, the fluorescence staining intensity of mutant HBsAg (mtHBsAg) was markedly weaker than that of the corresponding WT HBsAg, especially for the sW172* MT (Fig. 2), consistently demonstrating impaired expression. Compared to the even distribution of other HBsAgs in the cytoplasm, a more obvious dot-like distribution of mtHBsAg was observed in the cytoplasm of cells transfected with pHBV-sW196* compared to that transfected with pHBV-sW172* (Fig. 2), indicating intracellular retention of truncated HBsAg proteins. Therefore, the studied RT/surface truncation MTs impaired the expression and secretion of HBV surface proteins.
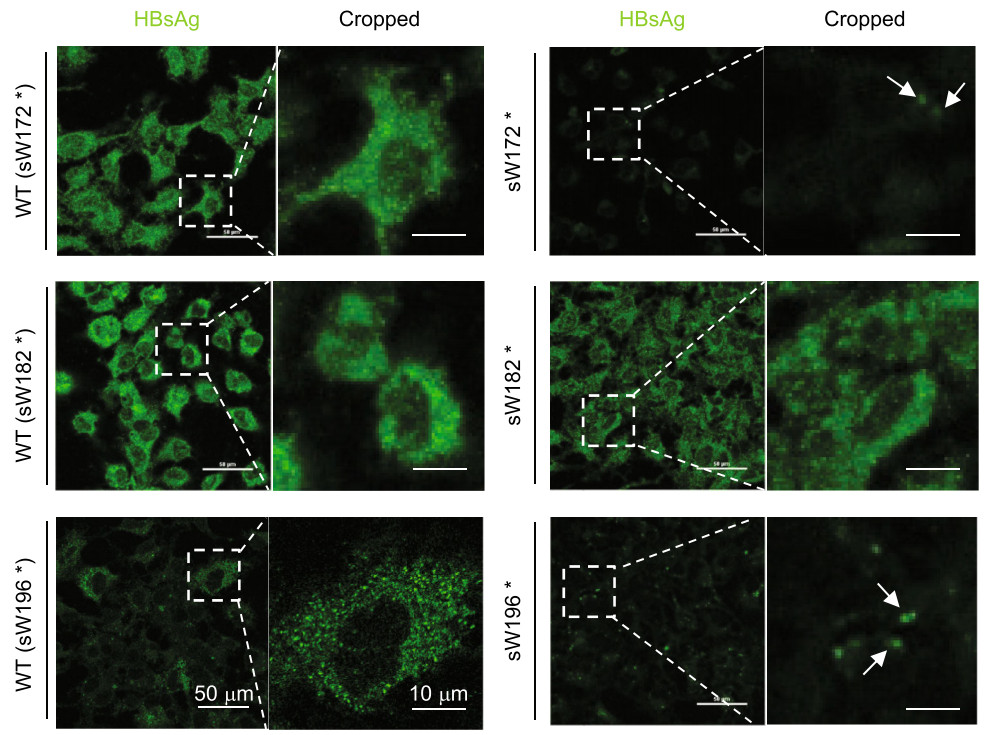
Figure 2. The rtM204I/sW196* mutation led to intracellular retention of HBsAg. Huh7 cells were seeded at approximately 30% confluence in 6-well plates and transfected with 1.5 μg pHBV-sW172* (sW172*), pHBV-sW182* (sW182*), or pHBV-sW196* (sW196*) and their corresponding plasmids containing WT HBV, pHBV-WT(sW172*) (WT(sW172*)), pHBV-WT(sW182*) (WT(sW182*)) or pHBVWT(sW196*) (WT(sW196*)), respectively for 48 h followed by plating on glass cover slips for 24 h. The cells were immunostained with an anti-HBs antibody S1 at 72 h posttransfection (hpt) (Scale bar: 50 μm). Higher magnification images of the selected area are also shown (Scale bar: 10 μm). The dot-like distribution of mtHBsAg is indicated by the white arrow.
-
The rtA181T/sW172* MT has been reported to have a secretory defect and exert a dominant-negative effect on WT HBV virion secretion (Warner and Locarnini 2008). Thus, we further analyzed the influence of sW182* and sW196* MTs on WT HBV virion secretion by cotransfecting a gradient concentration of MTs and their corresponding WT plasmids. Increasing amounts of pHBVsW172*, pHBV-sW182* and pHBV-sW196*, led to an increase in intracellular encapsidated DNA levels (Fig. 3A) but a decrease of DNA levels in secreted virions (Fig. 3B, 3C). In particular, a most profound decrease was observed when pHBV-sW172* or pHBV-sW182* were cotransfected with their corresponding WT plasmids at a ratio of 4 to 1, yielding an 80% decrease compared to pHBV-WT alone (Fig. 3B, 3C). In contrast, a less profound dosedependent decrease in HBsAg was observed in supernatants of both sW182*-WT (sW182*) and sW196*-WT (sW196*) cotransfected cells but not in supernatants of sW172*-WT (sW172*) cotransfected cells (Fig. 3D). These results demonstrated that the RT/surface truncation MTs had a dose-dependent negative effect on both WT HBV virion and HBsAg secretion in supernatants.
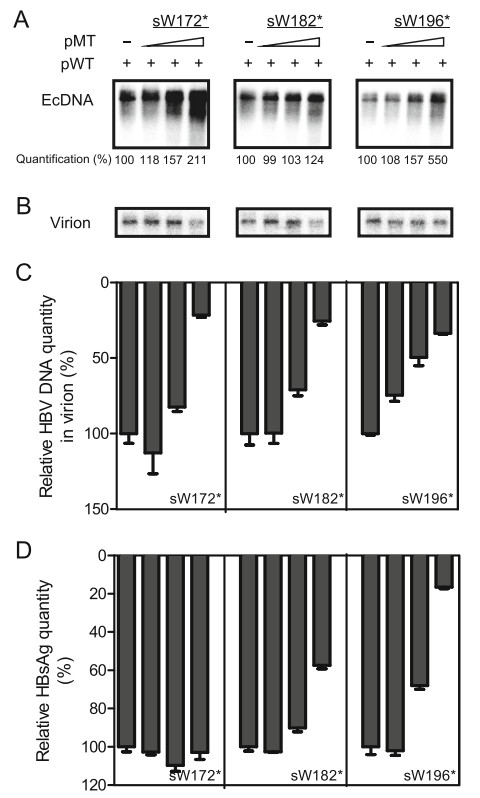
Figure 3. RT/surface truncation mutants had a dose-dependent negative effect on WT HBV virion secretion. Huh7 cells were seeded at approximately 30% confluence in 6-well plates and cotransfected with 0.4 μg of pHBV-WT(sW172*) (WT(sW172*)), pHBVWT(sW182*) (WT(sW182*)) or pHBV-WT(sW196*) (WT(sW196*)) and 0 lg, 0.1 lg, 0.4 μg or 1.6 μg of corresponding pHBV-sW172* (sW172*), pHBV-sW182* (sW182*), or pHBVsW196* (WT(sW196*)) plasmids, respectively. Cells were harvested at 72 h post transfection. A Encapsidated HBV DNA (EcDNA) was extracted from transfected cells and detected by Southern blotting. Numbers at the bottom indicate the relative band intensities for HBV replication intermediates, which were quantified using NIH ImageJ software. B–C Enveloped virions in culture supernatants were immunoprecipitated with anti-HBs antibodies. Virion-associated DNA was extracted and detected by Southern blotting (B) and realtime PCR (C). D HBsAg in cell culture supernatants was detected by ELISA. The relative quantity of HBV DNA in virions or HBsAg is presented as the percentage of DNA or protein in pHBV-WTtransfected samples alone.
-
NA resistance MTs are usually detected as a mixed population with WT HBV. However, it remains unknown how they affect each other, particularly under drug treatment. Therefore, the influence of RT/surface truncation MT and WT coexistence on viral replication competence was further analyzed by a cotransfection experiment at the indicated ratios (Fig. 4). Compare to the DNA level of WT HBV in the absence of LMV (Fig. 4A, Lane 1, Lane 6 and Lane 11), cotransfection of pHBV-WTs and pHBV-MTs at different ratios led to similar or even higher HBV DNA levels (Fig. 4A, Lane 4, Lanes 7–9 and Lanes 12–13). In particular, although the proportion of transfected MT plasmid increased, replication competence was not compromised (Fig. 4A, Lane 4, Lanes 8–9). Thus, coexistence of WT and MT HBV genomes maintained viral replication in the absence of LMV.
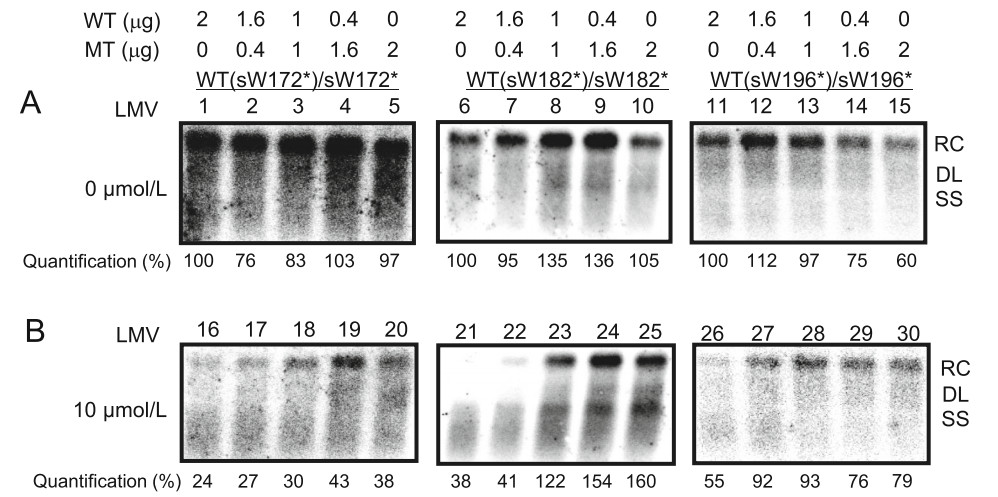
Figure 4. The presence of MTs maintained viral replication under LMV. Huh7 cells were seeded at approximately 30% confluence in 6-well plates and transfected with 2.0 μg pHBV-WTs [pHBV-WT(sW172*) (WT(sW172*)), pHBV-WT(sW182*) (WT(sW182*)) or pHBVWT(sW196*) (WT(sW196*))] or pHBV-MTs [pHBV-sW172* (sW172*), pHBV-sW182* (sW182*), or pHBV-sW196* (sW196*)] alone or cotransfected with pHBV-WTs and pHBV-MTs at indicated ratios and were harvested at 72 h post transfection. Intracellular HBV replication intermediates without (A) or with LMV treatment (B) were detected by Southern blotting. The positions of relaxed circular (RC), double stranded linear (DL), and single-stranded (SS) DNAs are indicated. Relative band intensities for HBV replication intermediates were quantified using NIH ImageJ software. The level of WT HBV DNA in the absence of LMV was considered the standard of replication, with this value set as 100%.
In contrast, LMV clearly inhibited replication of WT HBV (Fig. 4B, Lane 16, Lane 21 and Lane 26) but had no significant effect on replication of MT (sW182*) and MT (sW196*) HBV (Fig. 4B, Lane 25 and Lane 30), indicating a decreased susceptibility of MT HBV to LMV. When pHBV-WTs and pHBV-MTs were cotransfected, increasing amounts of pHBV-MTs led to an elevation in viral replication intermediates in the presence of LMV (Fig. 4B, Lanes 17–19, Lanes 22–24 and Lanes 27–29). Notably, HBV replication was rescued to a comparable level to that without LMV treatment when equal amounts of pHBVWT(sW182*) and pHBV-sW182* were cotransfected (Fig. 4B, Lane 23). These results indicated that the presence of MT HBV genomes maintained viral replication under LMV treatment.
-
As demonstrated in Fig. 3 and previous report (Warner and Locarnini 2008), RT/HBsAg stop codon MTs had a dominant-negative effect on WT HBV virion secretion. Therefore, we sought to determine the influence of the coexistence of MT and WT HBV genomes on virion production under LMV treatment. Compared with WT HBV, a similar amount of HBV DNA in virions was observed in the absence of LMV if WT and corresponding MTs were combined at a ratio of 4:1 and 1:1 (Fig. 5A, Lanes 2–3 to 1, Lanes 7–8 to 6, Lanes 12–13 to 11). However, a notably lower amount of HBV DNA was detected in virion fractions when WT and corresponding MT HBV genomes were mixed at a ratio of 1:4 (Fig. 5A, Lanes 4, Lane 9 and Lane 14), suggesting impaired secretion of HBV virions. We detected the replication of both WT and MT HBV under increasing concentrations of LMV. As shown in Supplementary Fig. S1, 10 μmol/L LMV significantly inhibited all WT HBV replication. In contrast, the susceptibility of all MT HBV to 10 μmol/L LMV was decreased. Similarly, 10 μmol/L LMV treatment significantly decreased the level of both intracellular and extracellular HBV DNA in virions when only pHBV-WTs were transfected (Fig. 5C, Lanes 16, Lane 21 and Lane 26 to Fig. 5A, Lanes 1, Lane 6 and Lane 11, respectively). Regardless, within the context of cotransfection of pHBV1.3-WTs and -MTs, intracellular HBV DNA was detected at a higher level (Fig. 5C, Lanes 17–18, Lanes 22–23 and Lanes 27–28), demonstrating that the presence of MT genomes maintained replication capacity under LMV. This result was also consistent with the findings shown in Fig. 4B. Correspondingly, the level of HBV DNA in virions was also higher (Fig. 5C, Lanes 17–18, Lanes 22–23 and Lanes 27–28). Moreover, when pHBV-WTs and -MTs were cotransfected at a ratio of 1:4, the level of HBV DNA in virions was significantly higher under LMV treatment than that without LMV treatment (Fig. 5C, Lanes 19, Lane 24 and Lane 29 to Fig. 5A, Lanes 4, Lane 9 and Lane 14, respectively), suggesting that impaired secretion of HBV virions without LMV treatment could be rescued under LMV treatment. In contrast, extracellular HBV DNA in virions was not detected due to the HBsAg secretion deficiency within the context of monotransfection of MTs (Fig. 5C, Lane 20, Lane 25 and Lane 30). Moreover, without LMV treatment, the level of HBsAg was decreased as the ratio of transfected WT plasmids to MT plasmids reduced (Fig. 5B). When LMV was added, HBsAg expression was only slightly decreased (Fig. 5D). This result also indicated that LMV treatment mainly inhibited WT replication but had only little effect on WT HBsAg expression. Overall, these results suggested that the presence of WT rescued the production of MT HBV virions under LMV treatment.
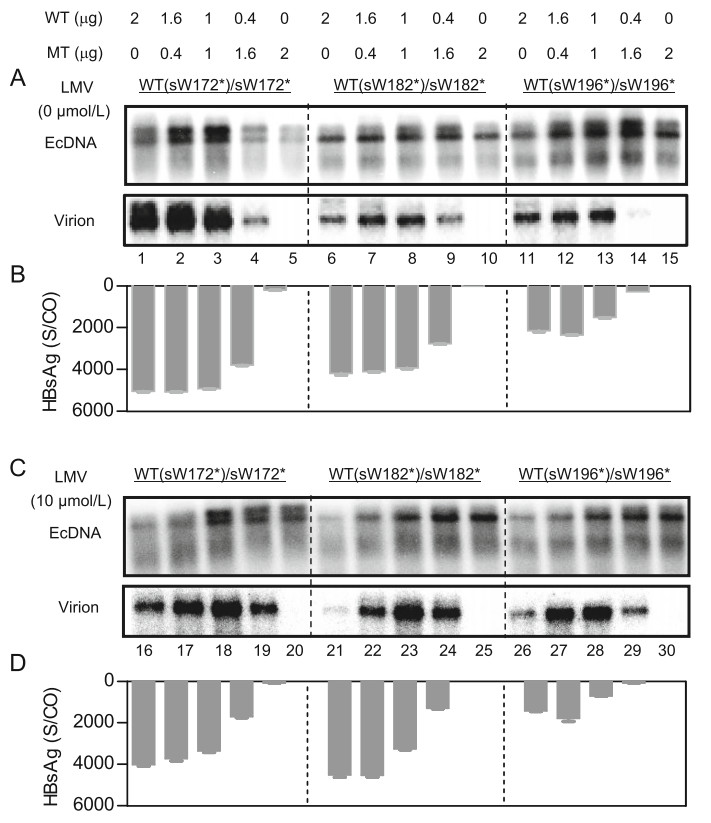
Figure 5. The presence of WT rescued production of MT HBV virions under lamivudine (LMV). Huh7 cells were seeded at approximately 30% confluence in 6-well plates and transfected with 2.0 μg pHBVWTs [pHBV-WT(sW172*) (WT(sW172*)), pHBVWT(sW182*) (WT(sW182*)) or pHBV-WT(sW196*) (WT(sW196*))] or pHBV-MTs [pHBV-sW172* (sW172*), pHBV-sW182* (sW182*), or pHBV-sW196* (sW196*)] alone or cotransfected pHBVWTs and pHBV-MTs at the indicated ratios and treated without (A, B) or with (C, D) LMV for 72 h. Encapsidated HBV DNA (EcDNA) was extracted from transfected cells or virionassociated DNA was extracted from immunoprecipitated enveloped virions in culture supernatants with anti-HBs antibodies and followed by Southern blotting (A, C). HBsAg in the cell culture medium were detected by ELISA (B, D).
The RT/surface Truncation MT Impaired Expression and Secretion of HBV Surface Proteins
RT/surface Truncation MTs Had a Dose-Dependent Negative Effect on WT HBV Virion Secretion
The Presence of MTs Maintained Viral Replication under lamivudine (LMV)
The Presence of WT Rescued the Production of MT HBV Virions under LMV
-
The emergence of RT/surface truncation MTs such as sW172* and sW182* in patients with chronic hepatitis B was previously reported to be associated with tumourigenicity (Lai and Yeh 2008; Lai et al. 2009; Yeh et al. 2011; Lee et al. 2012), which was suspected to be due to intracellular accumulation of mutated HBV surface proteins. In the present study, a dot-like distribution of HBsAg was observed in the cytoplasm of cells transfected with the pHBV-sW196* plasmid (Fig. 2), suggesting abnormal intracellular accumulation of HBsAg. Considering that accumulation of viral products in the endoplasmic reticulum (ER) of hepatocytes due to specific MTs such as the preS/S gene MTs may cause ER stress and hepatocyte injury (Pollicino et al. 2014), our data provide further evidence for the potential role of RT/surface truncation MT in tumorigenesis. Coincidently, a recent study reported that sW196* MT showed cytopathic effect (Colledge et al. 2017), which may be associated with the most obvious dotlike distribution within hepatoma cells observed for the sW196* MTs in this study (Fig. 2).
Previous observations have shown that the sW172* MT confer a dominant-negative effect with regard to HBsAg and viral particle secretion (Warner and Locarnini 2008; Ahn et al. 2014). Similar to this result, we demonstrated that in addition to sW172*, sW196* and sW182* MTs also impaired secretion of WT HBsAg and virions (Fig. 3). Compared with the results of Southern blotting (Fig. 3B), real-time PCR result uncovered a dose-dependent negative effect on the secretion of WT virions when different amounts of the sW196* or sW182* plasmid were cotransfected with the WT plasmid (Fig. 3C). Considering the difference in signal generation and detection between these two methods, Southern blotting and real-time PCR results may be not fully equal in numeric. Importantly, they show two complementary aspects of the results with the same tendency. In addition, compared to the secretion of virions, a less profound dose-dependent effect on HBsAg secretion was observed for cotransfection of the sW196* or sW182* plasmid with the WT HBV plasmid but not for sW172* with the WT HBV plasmid (Fig. 3). This result is consistent with previous reports (Colledge et al. 2017), presumably because the presence of WT HBV can partly rescue the secretion defect of truncated HBsAg. These results may be explained by the fact that the secretion pathway exploited by HBV subviral particles is quite different from that used by HBV virions (Lambert et al. 2007; Patient et al. 2007, 2009).
NA resistance MTs that result in a stop codon MT in HBsAg are usually detected as a mixed population with WT (Colledge et al. 2017), whereby RT/stop codon MTs may exert a dominant-negative effect on WT HBV virion secretion (Warner and Locarnini 2008). However, it remains unclear how the coexistence of RT/stop codon MTs and WT affects viral replication. In this study, we first found that the presence of WT HBV genomes rescued MT virion production under LMV treatment (Fig. 5). These results indicated that the existence of a low percentage of WT HBV under NA selective pressure complements the secretion deficiency of RT/stop codon MTs and allows HBV virion assembly and release of the defective MTs. In this manner, the defective MT HBV genomes could spread and persist stably in patients (Zoulim and Locarnini 2009).
In vitro phenotypic assays with a single HBV genome have been widely used to characterize the replication competence as well as the susceptibility of drug-resistant HBV genomes. Our findings suggest that these assays with a single HBV genome do not reflect the complex situation in patients. Therefore, phenotypic analysis of drug-resistant HBV genomes in combination with the respective WT HBV genome demands consideration.
-
We are grateful to Thekla Kemper (Institute of Virology, University Hospital of Essen, Essen, Germany), Barbara Bleekmann (Institute of Virology, University Hospital of Essen, Essen, Germany), Yuan Zhou (State Key Lab of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences), and Xue Hu (State Key Lab of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences) for excellent technical support. This work was supported by the Deutsche Forschungsgemeinschaft (TRR60), the National Nature Science Foundation of China (31770180, 31621061), the Youth Innovation Promotion Association CAS (No. 2016303), and the Youth Planning Project of Hubei Health Planning Commission (WJ2017Q027).
-
ML, XC and CW conceived and designed experiments. BL, XZ, KZ, YY and YC performed the experiments. CW, BL, XC and ML analyzed the data. YL, RC and DX contributed reagents/material/analysis tools. CW and ML wrote the manuscript.
-
The authors declare that they have no conflict of interest.
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: