HTML
-
The baculoviruses are a group of rod-shaped viruses with large DNA genomes that specifically infect insects. Two kinds of virion are produced during a typical baculovirus infection cycle: budded virus (BV) and occlusionderived virus (ODV), which mediates cell-to-cell and host-to-host infection, respectively (Keddie et al. 1989). Baculoviruses are phylogenetically divided into four genera, namely Alpha-, Beta-, Gamma-, and Deltabaculovirus (Jehle et al. 2006). The Alphabaculovirus is further divided into Group Ⅰ and Group Ⅱ based on phylogenetic analysis (Zanotto et al. 1993). Group Ⅰ and Ⅱ alphabaculoviruses are also characterized by using GP64 and ancestral F protein as their fusion proteins for BV, respectively (Pearson and Rohrmann 2002). Among all the baculoviruses, Group Ⅰ alphabaculoviruses are the most recently evolved (Herniou and Jehle 2007), and there are 12 specific genes that are only found in this lineage (Rohrmann 2011). The gp64 gene is one of them and has been suggested to be captured by an ancestral Group Ⅰ alphabaculovirus relatively late during evolution (Pearson and Rohrmann 2002). The rest 11 Group Ⅰ specific genes in the prototype baculovirus Autographa californica nucleopolyhedrovirus (AcMNPV) are: ac1 (ptp), ac5, ac16 (bv/odv-e26), ac27 (iap-1), ac30, ac72, ac73, ac114, ac124, ac132, and ac151 (ie2). It was proposed that the acquisition of the homologs of gp64 (ac128) and other Group Ⅰ specific genes may promote virus diversification and host range (Pearson et al. 2000; Herniou et al. 2001; Jiang et al. 2009).
To date, 10 of the 12 Group Ⅰ specific genes in AcMNPV have been studied. As mentioned above, gp64 encodes the viral major envelope fusion protein that essential for BV entry and infection (Monsma et al. 1996). The ac1 deletion can lead to partial defect in occlusion body (OB) formation in Spodoptera frugiperda 21 (Sf21) cells but not in Trichoplusia ni (T. ni) cells (TN-368 cells) (Li and Miller 1995). The ac5 encodes a protein that is an OB protein but not a component of BV or ODV, and is not required for BV production, the oral infectivity, and the formation of per os infectivity factor (PIF) complex (Wang et al. 2018). Inactivation of ac16 has no effects on protein synthesis in infected cells and oral infectivity to T. ni or S. frugiperda larvae (O'Reilly et al. 1990). ac27 is a gene of inhibiting apoptosis (Zeng et al. 2009), and its deletion appears to out-compete wild type virus in a cell-specific way (McLachlin et al. 2001). Deletion of ac30 has no obvious effects on BV production (Yu 2015). Though BV and ODV can be normally produced when ac114 or ac124 is deleted, the oral infectivity is significantly reduced or the time to kill infected larvae is increased, respectively (Wei et al. 2012; Liang et al. 2015). The product of ac132 is a nucleocapsid-associated protein essential for transport of nucleocapsid into nucleus (Fang et al. 2016). The ac151 seems to encode a protein that can facilitate DNA replication, virion formation, and infectivity in cell-specific way (Lu and Miller 1995; Prikhod'ko et al. 1999). Therefore, among the 10 studied Group Ⅰ specific genes of AcMNPV, only gp64 and ac132 are essential for virus infection, while others seem to contribute to virus infection in different aspects.
Currently, the function of ac72 and ac73 still remains unclear, although some studies on their homologs have been carried out in Bombyx mori nucleopolyhedrovirus (BmNPV). It was reported that the homolog of ac72 in BmNPV (bm58a) was not required for BV production, ODV assembly, or OB formation, however, it may function in promoting cell lysis and larval liquefaction (Yang et al. 2016). The bm59, a homolog to ac73, was first reported as an essential gene for virus infection (Ono et al. 2012), but was subsequently demonstrated to be dispensable for the propagation and assembly of BmNPV (Hu et al. 2016). Therefore, the reports on the role of bm59 during baculovirus life cycle seem to be controversial and its exact function remains to be clarified.
In this study, we aimed to characterize the function of ac73 during AcMNPV infection. We first detected the transcription and expression of ac73, and then studied the subcellular localization of AC73 during AcMNPV infection and determined whether it is a structure component of BV and ODV. The ac73 knockout and repaired recombinant viruses were constructed and characterized by in vitro and in vivo infection. Results showed that AC73 was expressed at late stage of virus infection and associated with the nucleocapsid fractions of both BV and ODV. Moreover, although ac73 is not an essential gene, it contributed to infectious BV production and viral infectivity in insect larvae to some extent.
-
Sf9 cells were cultured at 27 ℃ in Grace's insect medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS). The wild type (WT) AcMNPV E2 strain was obtained from the Microorganisms and Viruses Culture Collection Center, Wuhan Institute of Virology, Chinese Academy of Sciences (storage no. IVCAS1.0315). Ac-egfp and AcBac-egfp-ph were constructed previously (Wang et al. 2008; Shang et al. 2017). The AcMNPV bacmid (bMON14272) used for the construction of recombinant viruses was derived from the DH10BacTM Escherichia coli (E. coli) cells in Bac-to-Bac Baculovirus Expression System (Invitrogen, Carlsbad, CA, USA).
-
To generate specific polyclonal antibody against AC73 (anti-AC73), the open reading frame (ORF) of ac73 was amplified with 5'-GCGGAATTCATGAACACGTCCGTGGACG-3' (EcoRI site underlined) and 5'-GCGCTCGAGTTATTGTACATAATGTTTTATTGTAA-3' (XhoI site underlined) and inserted into the EcoRI and XhoI sites of pET-28a vector (Novagen, Carlsbad, CA, USA) to generate pET-28a-ac73. Then, the recombinant plasmid was electroporated into E. coli BL21 competent cells. The BL21 cells were induced with isopropyl-b-thiogalactopyranoside (IPTG) at 37 ℃ for protein expression. The expressed AC73 in BL21 cells was purified using cOmplete His-Tag Purification Resin (Roche Dnostics, Indianapolis, IN, USA) and the purified AC73 protein was used as antigen to generate rabbit polyclonal antiserum as previously described (Zou et al. 2016).
-
Sf9 cells were infected with WT AcMNPV at a multiplicity of infection (MOI) of 10 and harvested at 0, 3, 6, 12, 18, 24, 36, 48, and 72 h post infection (p.i.). For temporal transcription analysis, the total RNA of the infected cells was isolated by RNAiso Plus (TaKaRa, Dalian, China) according to the manufacturer's instruction. The DNA in RNA samples was eliminated and equal amounts (1 μg) of RNA were reverse transcribed to cDNA using the PrimeScriptTM RT reagent with the gDNA Eraser (TaKaRa, Dalian, China) Kit following the manufacturer's protocol. The total RNA or cDNA was used as the template for PCR amplification of an inner fragment of ac73 (primers: 5'-ATGAACACGTCCGTGGACG-3' and 5'-ACACCAATTTAAACACATGTTGAT-3'). To detect the expression of AC73 at different time points of infection, Sf9 cells were infected with WT AcMNPV using the same conditions above, and the cells were harvested and treated with protein sample buffer (50 mmol/L Tris-HCl, 2% SDS, 0.1% bromophenol blue, 10% glycerol, 5% 2-mercaptoethanol). Then, proteins were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose filter (NC) membrane (Millipore, Billerica, MA, USA) for Western blot analysis. The blots were incubated with anti-VP39 (Wang et al. 2010), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Li et al. 2018) or anti-AC73 antibody as the primary antibody, and HRP-conjugated goat anti-rabbit antibody (Sigma, St. Louis, MO, USA) as the secondary antibody. The bands were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL, USA).
-
For immunofluorescence analysis, about 1 × 106 Sf9 cells were infected with Ac-egfp virus at an MOI of 10, and then the cells were fixed at 12, 18, 24, 36, 48, and 72 h p.i. with 5% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for 10 min before being incubated with 5% BSA overnight at 4 ℃. Cells were then incubated with anti-AC73 antibody as the primary antibody at room temperature for 3 h and subsequently with Alexa 647- conjugated goat anti-rabbit (Abcam, Cambridge, UK) as the secondary antibody. The nuclei of infected cells were stained with Hoechst 33258 dye (Beyotime, Shanghai, China) for 5 min prior to the fluorescence microscopy. The fluorescence signals were observed by fluorescence microscopy (Deltavision softWoRx Imaging Workstation, Applied Precision).
-
To determine the localization of AC73 in transiently expressed cells, a plasmid containing egfp-fused ac73 was constructed as follows. First, the ORF of egfp was amplified from pEGFP-N1 (Clontech, Mountain View, CA, USA) and subcloned into the BamHI and EcoRI sites of pIZ/V5-His (Invitrogen, Carlsbad, CA, USA) to generate pIZ/V5-egfp. Then, the ORF of ac73 was amplified through PCR with primers 5'-GCGGAATTCATGAACACGTCCGTGGACG-3' (EcoRI site underlined) and 5'-GCGTCTAGATTATTGTACATAATGTTTTATTGTAA-3' (XbaI site underlined). Finally, the fragment was inserted into the EcoRI and XbaI sites of pIZ/V5-egfp to produce pIZ/V5-egfp-ac73. The pIZ/V5-egfp-ac73 plasmid was transfected into Sf9 cells using Cellfectin Ⅱ reagent (Gibco, Carlsbad, CA, USA). To detect the localization of EGFP fused AC73 in infected cells, Sf9 cells were transfected with pIZ/V5-egfp-ac73 plasmid as mentioned above and then infected with WT AcMNPV at an MOI of 5 for 48 h. Cells were then fixed and stained with Hoechst 33258 for observation as described above.
-
BVs from WT AcMNPV infected Sf9 cells and ODVs embedded in occlusion bodies (OBs) from virus infected larvae were purified as previously described (Braunagel and Summers 1994; Wang et al. 2010). The purified BVs and ODVs were further separated into envelope (E) and nucleocapsid (NC) fractions as previously described (Hou et al. 2013). Proteins in purified BVs, ODVs, and their E and NC fractions were then detected with anti-GP64, antiVP39, anti-ODV-E66 (Wang et al. 2010), anti-PIF5 (Wang et al. 2018), or anti-AC73 antibody by Western blots as described above.
-
A 326-base pair (bp) fragment upstream and a 220-bp fragment downstream of ac73 were PCR amplified with primers: AC73KO-UP-F (5'-TAAGGTACCCACGTTAGGCAGACAGTTG-3', KpnI site underlined) and AC73KO-UP-R (5'-GCGCTCGAGATATTTATTATTCCACGGACGTGTTCATG-3', XhoI site underlined) or AC73KO-Down-F (5'-GGGGATATCGCAACGCCATAG TGTTTGAC-3', EcoRV site underlined) and AC73KO-Down-R (5'-GGGTCTAGAGTGTCGCATCTAAGCGACG-3', XbaI site underlined). The two fragments were inserted into pKS-egfp-Cmr plasmid (provided by Dr. Just M. Vlak, Wageningen University and Research, the Netherlands) to generate pKS-ac73up-egfp-Cmr-ac73down plasmid. Then, the ac73up-egfp-Cmr-ac73down cassette was amplified through PCR using AC73KO-UP-F and AC73KO-Down-R primers. The purified linear fragment was electroporated into E. coli BW25113 competent cells containing AcMNPV bacmid (bMON14272) and λ Red recombinase-encoding plasmid pKD46 to generate AcΔ73 bacmid as described previously (Hou et al. 2002). A fragment, nucleotides (nt) 62757-62449, containing the promoter of ac73 was amplified from WT AcMNPV DNA with 5'-CAGCCCGGGCACGTTAGGCAGACAGTTG-3' (SmaI site underlined) and 5'-GGGCTCGAGGTTTCTTTTTTGAAAACTAAATTG-3' (XhoI site underlined). Then, the fragment was ligated into pFBD-ph (Li et al. 2018) to construct pFBD-Pac73-ph. The ORF of ac73 and the poly(A) signal sequence of ac73 were amplified with 5'-CGCCTCGAGATGTACCCATACGACGTCCCAGACTACGCTATGAACACGTCCGTGGACG-3' (XhoI site underlined; HAtag sequence in bold) and 5'-GGGGCATGCGTGTCGCATCTAAGCGACG-3' (SphI site underlined) and further inserted into pFBD-Pac73-ph to generate the donor plasmid pFBD-Pac73-HAtag-ac73-ph. The ac73 knockout (AcΔ73-ph) and repaired (AcΔ73-ac73R-ph) recombinant bacmids were generated by transposition of pFBD-ph or pFBD-Pac73-HAtag-ac73-ph into the LacZ mini-attTn7 locus of AcΔ73 using Bac-to-Bac Baculovirus Expression System (Invitrogen, Carlsbad, CA, USA).
-
About 1 × 106 Sf9 cells were transfected with recombinant bacmid DNA of AcΔ73-ph or AcΔ73-ac73R-ph with Cellfectin Ⅱ reagent, and fluorescence was observed at 48 and 96 h post transfection (p.t.) to determine the production of infectious BVs. At 120 h p.t., the supernatants were collected and used to infect healthy Sf9 cells for 48 h before fluorescence microscopy. To verify the correctness of the obtained AcΔ73-ph or AcΔ73-ac73R-ph virus, Sf9 cells were infected with AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph, then AC73 protein in the infected cells was detected through Western blot as described above. VP39 and GAPDH were also detected to serve as controls.
-
Cells were infected in triplicate with AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph virus at an MOI of 10, and 50 μL supernatant of each infection was collected at different time points post infection. The BV titers were determined by endpoint dilution assay (EPDA), and the averages of titers from three independent infections at each time point were calculated to plot one-step growth curves of these viruses. The statistical analysis was performed by one-way analysis of variance (ANOVA) method with SPSS software (IBM, Armonk, NY, USA).
-
For transmission electron microscopy (TEM) analysis, cells (1 × 106) were infected with AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph virus at an MOI of 10. At 24, 48, and 72 h p.i., cells were fixed with 2.5% glutaraldehyde. The samples were processed for TEM analysis as previously described (Li et al. 2018). The TEM images were taken using Tecnai G2 20 TWIN TEM (FEI, Hillsboro, OR, USA) at an accelerating voltage of 200 kV.
-
The bioassay was conducted using a droplet method (Hughes et al. 1986). Briefly, 48 early third-instar S. exigua larvae were fed with the OBs of AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph using droplet method at the concentration of 1 × 104, 3 × 104, 1 × 105, 3 × 105, 1 × 106, 3 × 106, 1 × 107, or 3 × 107 OBs/mL. Bioassays were performed twice and the infected larvae were monitored daily until all larvae had either pupated or died. The calculation of median lethal concentration (LC50) and the 95% confidence limits (CL) or the comparison of LC50 values among viruses were carried out using Probit regression method or the potency ratio test in SPSS software.
Cells and Viruses
Generation of Polyclonal Antibody against AC73
Time Course Analysis of ac73 Transcription and Expression
Immunofluorescence Microscopy of AC73
Localization Analysis of Fluorescent Protein Fused AC73
Detection of AC73 in BV and ODV
Construction of ac73 Knockout and Repaired Recombinant Bacmids
Production and Identification of Recombinant BVs
One-Step Growth Curve
Electron Microscopy (EM)
Bioassay
-
To study the transcription of ac73 in AcMNPV infected Sf9 cells, we first predicted the promoter of ac73. A TTAAG motif which is the typical feature of baculovirus late promoter (Morris and Miller 1994), was found from nt 61–57 upstream of ATG of ac73 (Fig. 1A), indicating that ac73 may be a late gene. This was consistent with a report that in AcMNPV infected T. ni cells, ac73 was mainly transcribed during late infection at nt 57 upstream of ATG of ac73 (Chen et al. 2013). To further determine whether ac73 is really a late gene, the transcripts of ac73 at different time points of infection were detected through PCR amplification of an inner fragment (~ 270 bp) within ac73. Result showed that the transcripts of ac73 could be detected from 12 to 72 h p.i. (Fig. 1B), indicating that ac73 was expressed at the late stage of infection. In addition, Western blot analysis was performed to detect AC73 protein levels in infected cells. The AC73 protein was under the detectable level before 18 h p.i., but was clearly detected since 24 h p.i. (Fig. 1C). For reference, VP39, the well-known viral late protein (Thiem and Miller 1989), could also be clearly detected since 18 h p.i. (Fig. 1C). With the above results, we can conclude that ac73 is a late gene of AcMNPV.
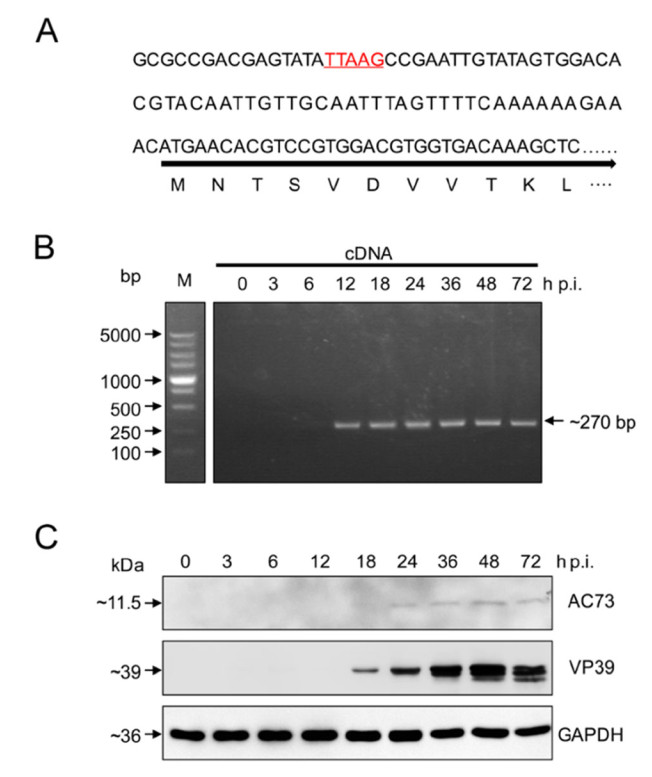
Figure 1. Ac73 is a late viral gene. A The promoter prediction of ac73. The predicted late transcription motif TTAAG is underlined and indicated in red. B Transcription detection of ac73. The ac73 transcripts from different time points of infection were detected through PCR amplification of a fragment of the ORF ac73 (~ 270 bp) using cDNA as templates. bp, base pair. C Expression detection of ac73. The AC73 protein at different time points of infection was detected by Western blot using anti-AC73 antibody. VP39 which is expressed during late infection served as a control together with cellular GAPDH.
-
Next, the subcellular localization of AC73 in infected cells was determined by immunofluorescence microscopy. In Ac-egfp infected cells, AC73 could be clearly detected in the nucleus of infected cells since 18 h p.i., and it was mainly localized in the ring zone region peripheral to the nuclear membrane during virus infection (Fig. 2A). To further determine whether AC73 can enter the nucleus independently, EGFP fused AC73 or the control EGFP alone was transiently expressed by transfection of the corresponding plasmid into Sf9 cells. The result showed that both EGFP and EGFP fused AC73 were evenly distributed in the cytoplasm and nucleus (Fig. 2B), suggesting that AC73 alone could not enter the nucleus completely. This is consistent with the fact that no nuclear localization signal could be predicted in AC73 (data not shown). To exclude the possibility of effect of EGFP on the localization of AC73, Sf9 cells were first transfected with plasmid encoding EGFP or EGFP fused AC73 and then infected with WT AcMNPV. Compared to the result of transient expression, EGFP fused AC73 but not EGFP in the superinfected cells showed clear nuclear localization and was embedded into OBs (Fig. 2B). Thus, the results suggested that AC73 could enter the nucleus in an infectiondependent way and it seemed to be assembled into OBs.
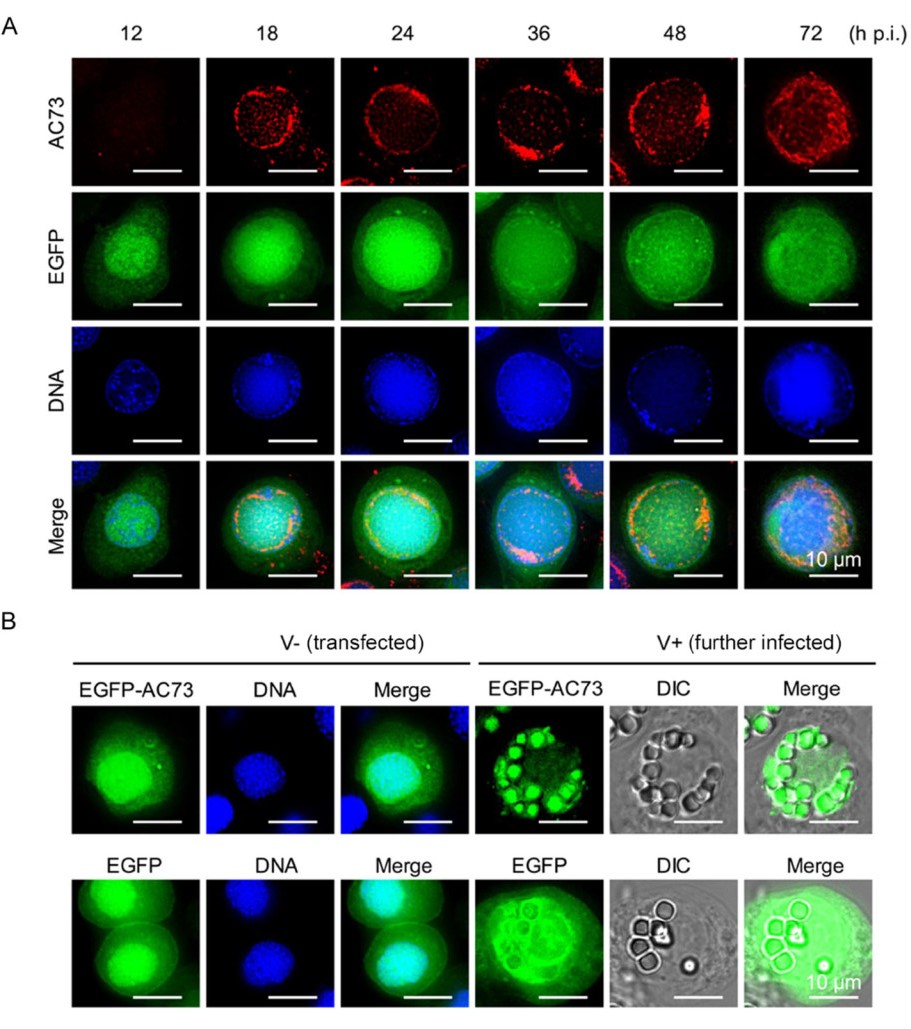
Figure 2. The subcellular localization of AC73. A Immunofluorescence assay of AC73 localization during Acegfp infection. Sf9 cells were infected with Ac-egfp which does not contain ph, thus no OB formed. Cells were fixed at indicated time points of infection, and the anti-AC73 antibody and Alexa 647-conjugated goat anti-rabbit antibody were used as the primary and secondary antibody for detection of AC73, respectively. EGFP fluorescence indicated the cells were successfully infected; the nuclei of cells were stained with Hoechst 33258 dye (blue). B The localization of EGFP fused AC73 in transfected and infected cells. V - the cells were transfected with plasmid encoding EGFP or EGFPAC73. V + transfected cells (V -) were further infected with WT AcMNPV which can express polyhedrin for the formation of OBs and observed at 48 h p.i.. DIC differential interference contrast. Bars, 10 μm.
-
Previous proteomics data revealed that AC73 was associated with BV (Wang et al. 2010), but not with ODV (Braunagel et al. 2003). However, our result found that EGFP fused AC73 could be assembled into OBs, suggesting that AC73 may also be ODV-associated. To test this possibility, BVs and ODVs were prepared from the supernatant of WT AcMNPV infected cells and larvae, respectively, and then analyzed by Western blot with antiAC73 antibody. As shown in Fig. 3A, AC73 could be probed in both BV and ODV samples. To further confirm this result and to determine the localization of AC73 in virion more accurately, the BVs and ODVs were fractionated into envelope and nucleocapsid components. As the Western blot result showed, AC73 could be detected in nucleocapsid samples of BV and ODV, but not in the envelope samples (Fig. 3B). Thus, AC73 is a nucleocapsid component of both BV and ODV.
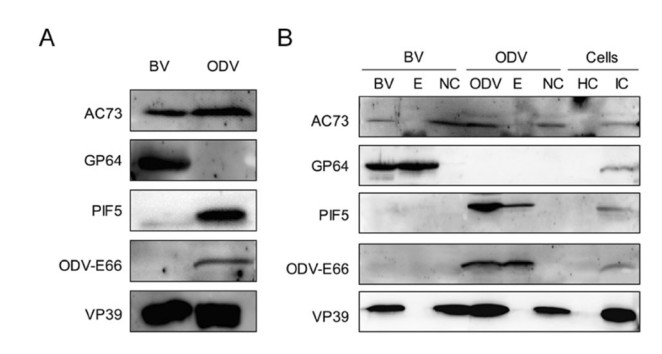
Figure 3. AC73 is a nucleocapsid protein of BV and ODV. A AC73 is the component of BV and ODV. The BVs and ODVs were purified and the intact virions were subjected to SDS-PAGE for Western blot assay by using anti-AC73. B AC73 is a nucleocapsid protein. The purified BVs and ODVs were fractionated into envelope (E) and nucleocapsid (NC) fractions, and then analyzed together with intact BV and ODV samples. The healthy cells (HC) and infected cells (IC) were served as controls. for A, B GP64 serves as positive control for the envelope of BV; PIF5 and ODV-E66 served as positive controls for the envelope of ODV; VP39 was the control for nucleocapsid of BV and ODV.
-
To determine the function of AC73 in virus infection, ac73 knockout and repaired bacmids were constructed. A 168 bp of ac73 in AcMNPV bacmid bMON14272 was replaced with egfp and Cmr genes to generate AcΔ73 bacmid, and then ph alone or both ph and HAtag-fused ac73 genes were inserted into the AcΔ73 bacmid to produce AcΔ73-ph or AcΔ73-ac73R-ph bacmid (Fig. 4A). Then, AcΔ73-ph or AcΔ73-ac73R-ph bacmid was transfected into Sf9 cells. The result showed that the number of fluorescent cells increased obviously from 48 to 96 h p.t. in both AcΔ73-ph and AcΔ73-ac73R-ph transfected cells (Fig. 4B, left two panels), indicating that infectious BVs could be produced when ac73 was deleted. To further confirm this, the supernatants from transfected cells were collected and then used to infect healthy Sf9 cells. Cells were successfully infected with AcΔ73-ph or AcΔ73-ac73R-ph virus as indicated by the occurrence of EGFP fluorescence (Fig. 4B, right panel). To confirm the correctness of recombinant viruses, AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph virus infected Sf9 cells were analyzed by Western blot. The result showed that the WT AC73 and HAtag-AC73 protein could be detected in AcBac-egfp-ph and AcΔ73-ac73R-ph infected cells, respectively, but no signal could be detected in AcΔ73-ph infected cells (Fig. 4C), suggesting AcΔ73-ph and AcΔ73-ac73R-ph were correctly constructed and produced. Taking together, these results indicated that ac73 is non-essential for BV propagation in cultured cells.
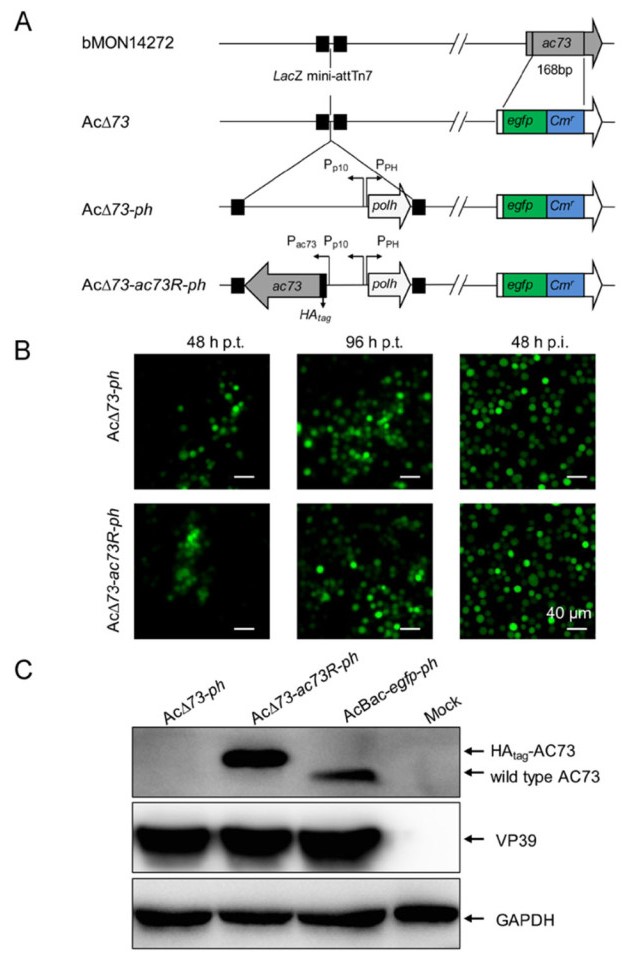
Figure 4. Construction and identification of ac73 knockout and repaired viruses. A Construction of AC73 knockout and repaired bacmids. A fragment of ac73 in AcMNPV bacmid was replaced by egfp and Cmr, and was then inserted with ph or both ph and ac73 to construct AC73 knockout (AcΔ73-ph) or repaired (AcΔ73-ac73R-ph) bacmid. B Transfection and infection assay. Sf9 cells were transfected with the bacmid of AcΔ73-ph or AcΔ73-ac73R-ph and observed at 48 and 96 h p.t. (the left two panels). At 120 h p.t., the supernatants of infected cells were collected and used to infect healthy Sf9 cells, and the infections were detected at 48 h p.i. based on fluorescence (the right panel). Bars, 40 μm. C Detection of AC73 in the recombinant viruses infected cells. Sf9 cells were infected with AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph virus at an MOI of 10 for 36 h and then collected for Western blot assay. VP39 and GAPDH were used as controls.
-
To quantify whether ac73 contributes to BV production, one-step growth curve analysis of AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph was performed. Sf9 cells were infected with these viruses at an MOI of 10, and BV titers at 0, 24, 48, 72, and 96 h p.i. were determined. In contrast to the result of bm59 deletion which did not affect BV production (Hu et al. 2016), one-step growth curve assay revealed the BV titers of AcΔ73-ph decreased by approximately 8- and 5-fold compared to that of AcBac-egfp-ph virus at 72 and 96 h p.i. respectively (P < 0.05) (Fig. 5). By comparison, at all the time points of the infection, the BV titers of AcΔ73-ac73R-ph showed no significant difference with those of AcBac-egfp-ph (P> 0.05) (Fig. 5). Thus, though ac73 is non-essential for BV production, it does play a role in optimal production of infectious BVs.
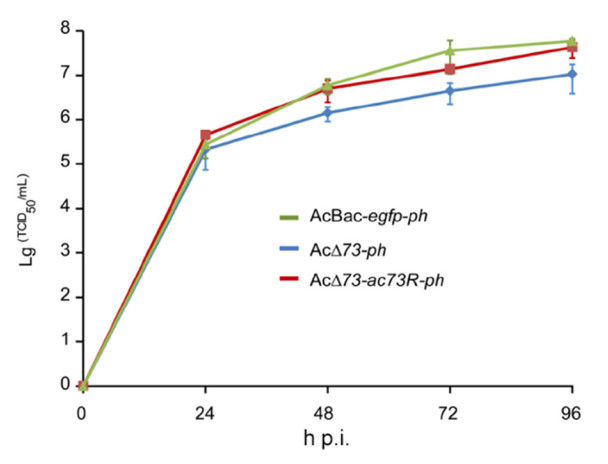
Figure 5. Deletion of ac73 decreased infectious BV production. Sf9 cells were infected with AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph virus at an MOI of 10. The supernatants of infected cells were harvested at the indicated time points of infection, and virus titers were determined by endpoint dilution assay for one-step growth curve analysis. The points represent the average titers from triplicate infections and error bars indicate standard deviations (SD).
-
Next, we determined whether ac73 is essential for the morphogenesis of ODV and OB. To this end, AcBac-egfpph, AcΔ73-ph, and AcΔ73-ac73R-ph infected cells at 24, 48, and 72 h p.i. were subjected to electron microscopy. At 24 h p.i., the nucleocapsids and ODVs could be detected in the nucleus of infected cells for AcBac-egfp-ph and AcΔ73-ac73R-ph, as well as AcΔ73-ph (Fig. 6, upper panel), suggesting the ac73 was neither essential for the nucleocapsid assembly, nor for the envelopment of nucleocapsids to form ODV. At 48 and 72 h p.i., the OBs that embedded with ODVs were formed in AcBac-egfp-ph and AcΔ73-ac73R-ph, as well as in AcΔ73-ph infected cells (Fig. 6, lower two panels). These results showed that ac73 was not required for ODV or OB formation in infected cells.
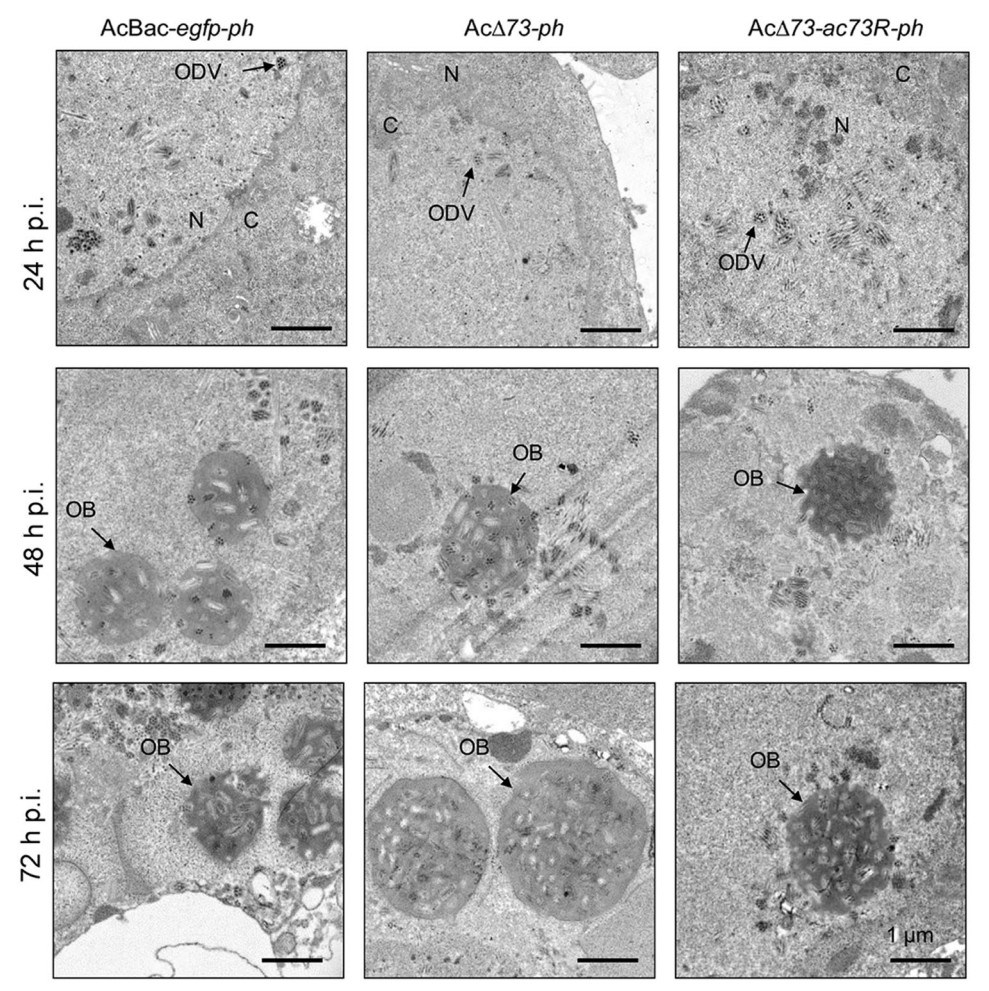
Figure 6. EM analysis of ODV and OB formation. Sf9 cells were infected with AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph virus at an MOI of 10 and fixed at indicated time points of infection for EM analysis. The representative ODVs or OBs are indicated by black arrow. N nucleus, C cytoplasm. Bars, 1 μm.
-
To further investigate the function of AC73 in vivo, bioassay was performed to determine the effects of ac73 deletion on viral infectivity in host level. The early thirdinstar S. exigua larvae were orally infected with OBs of AcBac-egfp-ph, AcΔ73-ph, or AcΔ73-ac73R-ph at different concentrations using droplet method (Hughes et al. 1986). Liquefaction of the infected larvae after death was observed for AcBac-egfp-ph, AcΔ73-ph, and AcΔ73-ac73R-ph viruses (data not shown), indicating that ac73 was not essential for oral infection and liquefaction of infected larvae. In two independent experiments, the potency ratio test showed that there was no significant difference between the AcBac-egfp-ph and AcΔ73-ac73Rph viruses as evidenced by the including of the value 1.0 for 95% CL (Robertson et al. 2007), but the LC50 of AcΔ73-ph virus was 3–4 fold higher than that of AcBac-egfp-ph virus (the potency ratio didn't include 1.0) (Table 1), suggesting that the deletion of ac73 reduced the viral infectivity of AcMNPV in S. exigua larvae. Thus, ac73 is a virulent gene that contributes to virus infection in vivo.
Virus Test 1 Test 2 LC50(95% CL)
(× 104 OBs/mL)Potency ratioa
(95% CL)LC50 (95% CL)
(× 104 OBs/mL)Potency ratioa(95% CL) AcBac-egfp-ph 6.84 (4.35, 10.53) 5.19 (2.77, 9.26) AcΔ73-ph 25.32 (16.60, 38.30) 3.70 (1.96, 7.56) 15.14 (8.71, 25.57) 2.92 (1.312, 7.20) AcΔ73-ac73R-ph 6.71 (4.24, 10.40) 0.98 (0.53, 1.82) 2.79 (1.43, 5.12) 0.538 (0.22, 1.24) aThe potency ratio was calculated by dividing the LC50 of the AcΔ73-ph and AcΔ73-ac73R-ph viruses by that of AcBac-egfp-ph. A significant difference was based on whether the 95% confidence limits (CL) of the potency ratio included the value 1.0. Table 1. Bioassay results of recombinant viruses against early third-instar S. exigua larvae.
Ac73 Is a Late Viral Gene
Cellular Localization of AC73
AC73 is a Nucleocapsid Protein of Both BV and ODV
Ac73 is Not Essential for Infectious BV Production
The ac73 Deletion Resulted in Decreased Production of Infectious BVs
Electron Microscopy of AcBac-egfp-ph, AcΔ73-ph, and AcΔ73-ac73R-ph infected cells
The Effects of ac73 Deletion on Viral Infectivity in Insect Larvae
-
The ac73 is one of the 12 specific genes of Group Ⅰ alphabaculoviruses, but its function during AcMNPV infection was unknown. In this study, we showed that ac73 was a late gene (Fig. 1) and its product, AC73, was associated with the nucleocapsid fractions of both BV and ODV (Fig. 3). In addition, AC73 appeared to be assembled into the OBs (Fig. 2B). Deletion of ac73 resulted in about 5–8 fold decrease of BV production at late stages of viral infection (Fig. 5) and about 3–4 fold decrease of viral infectivity in host level (Table 1). Therefore, like many of other Group Ⅰ specific proteins which are not essential but may benefit for virus infection, such as AC1 (Li and Miller 1995), AC114 (Wei et al. 2012), and AC124 (Liang et al. 2015), AC73 is also a luxury protein that remained/captured during evolution to contribute to virus infection.
Some of our results of ac73 were different from the studies of its homologue bm59 in BmNPV. First, ac73 was found as a late gene and this was consistent with the present of a late transcription motif TTAAG in its promoter region (Fig. 1A) as well as the transcriptome result of AcMNPV in T. ni cells (Morris and Miller 1994; Chen et al. 2013). However, bm59 was characterized as an early gene with an atypical transcriptional start motif, CAAC motif (Hu et al. 2016). We found that a TTAAG motif is also present at nt 61-57 upstream of the ATG of bm59. It remains unknown why bm59 does not use the conserved TTAAG motif for late gene transcription. Second, deletion of ac73 resulted in the reduction of infectious BV production, however, bm59 deletion was initially reported to be essential for BV production (Ono et al. 2012), but later showed no impact on infectious BV production (Hu et al. 2016). Considering AC73 and Bm59 share high sequence similarity (~ 90% aa identity), these differences may reflect that ac73 and bm59 are diverged at relatively late stage of baculovirus evolution and are adapting to their different hosts.
Actually, it is not surprising to find that the Group Ⅰ specific gene homologues function differently in AcMNPV and BmNPV. For example, ac5 had no obvious effects on OB formation when deleted (Wang et al. 2018), but its homolog, bm134, was found to be important for the embedding of ODVs into OBs in BmNPV (Shen et al. 2018). In addition, though ac16 deleted virus could produce infectious BVs (O'Reilly et al. 1990), its homolog, bm8, is essential for infectious BV production (Imai et al. 2004). Similarly, BVs could be normally produced when ac124 was deleted (Liang et al. 2015), but bm101 (homolog of ac124) was found to be essential for BV production in BmNPV (Chen et al. 2014). Although AcMNPV and BmNPV are two closely related viruses with an average ~ 90% amino acid sequence identity between homologous ORFs (Gomi et al. 1999), they show a striking difference in host range. BmNPV is host specific that it only infects B. mori or B. mandarina (Shirata et al. 1999; Iwanaga et al. 2009), but AcMNPV shows a wide host range of at least 32 lepidopteran insect species (Granados and Williams 1986), yet it is unable to complete a productive replication in B. mori cells or kill B. mori larvae (Morris and Miller 1993; Grasela et al. 2000). In contrast to the functional diversity between the Group Ⅰ specific genes of the two viruses, the function of most other genes appeared to be relatively consistent between AcMNPV and BmNPV (data not shown). Therefore, our study and previous studies highlighted the uniqueness of Group Ⅰ specific genes in the evolution of alphabaculoviruses.
When searched against non-redundant protein database at National Center for Biotechnology Information (NCBI) using Position-Specific Iterated Basic Local Alignment Search Tool (PSI-BLAST), AC73 was found to share high similarity with Bcl-2-associated athanogene (BAG) domains of some proteins, for example, the BAG domain of Starvin protein from Drosophila melanogaster which is required for larval food uptake and involved in the recovery from cold stress (Coulson et al. 2005; Colinet and Hoffmann 2010), the BAG domain of Samui protein from B. mori which is cold-inducible and can protect against cold-injures or transmit cold signal for gene expression (Moribe et al. 2001), and the BAG domain of BAG-4 (also known as silencer of death domains (SODD)) from Homo sapiens which interacts with Hsp70 or tumor necrosis factor receptor type 1 (TNFR1) to affect cell death (Miki and Eddy 2002). A BAG domain can bind to the ATPase domain of Hsc70/Hsp70 to regulate its activity (Bimston et al. 1998; Terada and Mori 2000; Gassler et al. 2001). Therefore, ac73 may be acquired from a host during evolution and produce a protein to mimic the functions of host BAG domain-containing proteins to facilitate virus infection under certain conditions. But further investigations are required to identify whether AC73 functions as a BAG domain-like protein and to unravel the detailed role and the function mode of ac73 in virus life cycle.
-
This research was supported by the grants from the Key Research Project of Frontier Science (QYZDJ-SSWSMC021), the Strategic Priority Research Program (grant No. XDB11030400) from the Chinese Academy of Sciences, and the grants (No. 31621061) from the National Natural Science Foundation of China. We would like to thank Mr. Ding Gao, Ms. Pei Zhang and Ms. An-na Du, Mr. He Zhao, and Ms. Li Li from The Core Facility and Technical Support of Wuhan Institute of Virology for their technical assistance.
-
WS, ZH and MW designed the experiments. WS, LH and QC performed the experiments and analyzed the data. WS, ZH and MW wrote the manuscript. JL, FD, HW, ZH and MW edited and commented on the manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: