-
Abbreviations ARRDC3 Arrestin domain containing 3 BRE Brain and reproductive organ-expressed CCRN4L Carbon catabolite repression 4-like CDCA3 Cell division cycle associated 3 CVB3 Coxsackievirus B3 FIBP FGF1 intracellular binding protein HLA-DQA1 Major histocompatibility complex, class Ⅱ, DQ alpha 1 hnRNPH3 Heterogeneous nuclear ribonucleoprotein H3 IRF2BP1 Insulin-like growth factor 2 mRNA-binding protein 1 KEGG Kyoto Encyclopedia of Genes and Genomes LncRNA Long non-coding RNA MDA5 Melanoma differentiation associated gene 5 MTX1 Metaxin 1 NEMF Nuclear export mediator factor NFAT5 Nuclear factor of activated T cells 5 SNHG11 Small nucleolar RNA host gene 11 TIMP1 TIMP metallopeptidase inhibitor 1 TMED9 Transmembrane p24 trafficking protein 9 ZNF295 Zinc finger protein 295
HTML
-
Long noncoding RNAs (lncRNAs) are classically defined as RNA transcripts larger than 200 nucleotides without any protein-coding capacity (Cheetham et al. 2013; Ma et al. 2013), although recent publications have reported that several lncRNAs retain limited translational activity and code small peptides (Anderson et al. 2015; Matsumoto et al. 2017). In cells, lncRNAs play various regulatory roles via interacting with DNA, RNA or protein molecules, modulating gene expression and protein functions (Ponting et al. 2009; Benetatos et al. 2011; Fatica and Bozzoni 2014). Recently, accumulated studies demonstrate critical roles of host lncRNAs in viral infection (Liu and Ding 2017). For example, lnc-Lsm3b is up-regulated upon the infection of different viruses such as vesicular stomatitis virus, sendai virus and herpes simplex virus, inhibits the stimulation of IFN-α and -β, and thus blocks cellular antiviral response (Jiang et al. 2018). By contrast, lncITPRIP-1 induced by the infection of hepatitis C virus mediates MDA5-triggered production of IFNs and ISGs, leading to the suppression of HCV replication (Xie et al. 2018).
Coxsackievirus B3 (CVB3), a pathogenic enterovirus of Picornaviridae family, is a common causative agent of various inflammatory diseases including myocarditis (Zhang et al. 1997; Fairweather et al. 2012), meningitis (Wong et al. 2011), pancreatitis (Ursing 1973; Mena et al. 2000) and hepatitis (Liu et al. 2013). Especially, CVB3- induced myocarditis frequently progresses to dilated cardiomyopathy (DCM), which is a major cause of sudden unexpected death in patients less than 40 years of age (Kearney et al. 2001; Fung et al. 2016). Currently, there is no effective therapy available for acute CVB3 infection, and as for DCM, the only treatment is heart transplantation (Kearney et al. 2001). Therefore, it is critical to identify novel drug targets for therapy development of CVB3-related diseases.
During the life cycle of CVB3, the virus interacts with various cellular components of the host, which either benefits viral survival or is a defensive mechanism of host to counteract viral infection (Zhai et al. 2018). For instance, CVB3 promotes the expression of a molecular chaperone protein Hsp70 that stabilizes viral genomic RNA and enhances viral translation (Qiu et al. 2016; Wang et al. 2017); CVB3 infection induces the up-regulation of a stress-responsive protein NFAT5 which inhibits CVB3 replication, but this antiviral protein is cleaved and inactivated by CVB3 proteases at later infectious stage (Qiu et al. 2017). In addition to host proteins, CVB3 also interplays with cellular non-coding RNAs, especially microRNAs. Our previous studies demonstrated that miR- 342-5p suppresses CVB3 biogenesis via targeting the coding region of the viral protein 2C (Wang et al. 2012), while miR-10a* favors CVB3 biosynthesis via targeting the viral-3D-coding region (Tong et al. 2013). However, the interactions between CVB3 and lncRNAs have not been revealed to date, although lncRNAs are also closely related to several pathways critical for CVB3 replication.
To uncover the roles of lncRNA in CVB3 infection, we employed mRNA function analyses and the correlation network of lncRNA-mRNA expression. Firstly, we screened the expression profiles of lncRNAs and mRNAs in CVB3-infected and -uninfected HeLa cells by microarray analysis. Based on the data, we summarized the function of all differentially expressed mRNAs by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. Finally, we established the correlation network of differentially expressed lncRNAs and mRNAs, which facilitated the elucidation of the lncRNA-mRNA interactions and the functional roles of lncRNAs in CVB3 infection.
-
HeLa cells (ATCC) were cultured using Dulbecco's Modified Eagle's Medium (DMEM, Gibco) supplemented with 100 μg/mL penicillin–streptomycin and 10% fetal bovine serum (FBS, Sigma) in 37 ℃ supplied with 5% of CO2.
CVB3 (CG) strain was a kind gift from Dr. Charles Gauntt (University of Texas Health Science Center) and propagated in HeLa cells. Virus stock was prepared from the cells by three freeze–thaw cycles followed by centrifugation to remove cell debris and stored at - 80 ℃. The titer of virus stock was determined by plaque assay as described below. Cell cultures were infected with CVB3 at 0.5 MOI for 1 h in serum-free medium, washed with phosphate-buffered saline (PBS, Thermo Fisher), and then replenished with fresh medium containing fetal bovine serum (FBS) followed by 23 h culture. Cells treated with the same amount of PBS in the same process served as uninfected control.
-
Total RNA was extracted and purified using TRIzolTM Plus RNA Purification Kit (Invitrogen). Total RNA then was reverse-transcribed to cDNA using SuperScriptTM Ⅳ FirstStrand Synthesis System (Invitrogen) according to the manufacturer's instructions.
-
Total RNA from each sample was quantified by the NanoDrop ND-1000 and RNA integrity was assessed by standard denaturing agarose gel electrophoresis. For microarray analysis, Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer's standard protocols with minor modifications. Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLYTM Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method. The labeled cRNAs were hybridized onto the Human LncRNA Array v3.0 (8 × 60K, Arraystar). After having washed the slides, the arrays were scanned by using the Agilent Scanner G2505C.
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Raw signal intensities were normalized in quantile method and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). Expression of GAPDH (glyceraldehyde phosphate dehydrogenase) was used as an internal control. LncRNAs or mRNAs with at least two-fold changes in the expression levels and FDR (false discovery rate) < 0.05 were deemed to be differentially expressed. The differentially expressed lncRNAs and mRNAs with statistical significance between two groups were identified through Volcano Plot filtering. Pathway analysis was applied to determine the roles of these differentially expressed mRNAs played in these biological pathways. Finally, Hierarchical clustering was performed to show the distinguishable lncRNAs and mRNAs expression pattern among samples.
-
Three siRNAs targeting XLOC_001188 were designed by the supplier (Suzhou GenePharma), among which only one effectively knocked down XLOC_001188 and was chosen to conduct the following experiments (Sense: 5′-GGUGAGUCUGGAAAGGCUUTT-3′; Antisense: 5′- AAGCCUUUCCAGACUCACCTT-3′). The siRNA was transfected to HeLa cells using Lipofectamine 2000 per the manufacturer protocol. At 48 h post transfection, the cells were subjected to CVB3 infection.
-
qPCR was performed in triplicate to validate the expression of significantly altered lncRNAs and mRNAs in CVB3-infected HeLa cells compared to uninfected control cells. Quantitative PCR analysis and data collection were performed on the ViiATM 7 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using the primer pairs listed (Supplementary Table S1). GAPDH served as an endogenous control for normalization. For relative quantification, 2-ΔΔCt was calculated and used as an indication of gene relative expression.
-
Pathway analysis was conducted according to Kyoto Encyclopedia of Genes and Genomes (KEGG: http://www.genome.ad.jp/kegg/), two-sided Fisher's exact test and χ2 test were used to select significant pathways, and the False Discovery Rate (FDR) was calculated to correct the P value (Draghici et al. 2007).
-
The lncRNA-mRNA correlation network was constructed by correlation calculation based on the values of the normalized signal intensity of specific expression in the differentially expressed genes. For each pair of mRNA-lncRNA, Pearson correlation was calculated to determine the significance of correlation and the correlation value cutoff was 0.92. Correlation degrees of lncRNAs and mRNAs were calculated by counting their correlated counterparts. The P-value denoted the significant level of gene co-expression and the threshold of significance was P-value < 0.05.
-
SPSS 20.0 software (SPSS Inc., Chicago, IL, USA) was used to do statistical analysis. All data were shown as mean ± SD of three independent experiments with each experiment in triplicate. The student t test was used to evaluate the expression differences of lncRNAs and mRNAs between CVB3-infected cells and uninfected control cells. P < 0.05 was considered as statistically significant.
Cell Culture and CVB3 Infection
RNA Preparation
Microarray Processing and Analysis
siRNA Transfection
Quantitative Real-Time PCR (qPCR)
Pathway Analysis
LncRNA-mRNA Correlation Network
Statistical Analysis
-
By microarray quantifying host lncRNAs and mRNAs in CVB3-infected HeLa cells and uninfected controls, we found 700 differentially expressed lncRNAs (Fig. 1A, Supplementary Table S2) and 665 differentially expressed mRNAs (Fig. 1B, Supplementary Table S3). Among the differentially expressed lncRNAs, 431 were up-regulated and 269 were down-regulated upon CVB3 infection; as for mRNAs, 299 were up-regulated and 366 were down-regulated (fold change > 2.0, P < 0.05). To evaluate the reliability of microarray results, we randomly selected five up-regulated and three down-regulated lncRNAs, and verified their expression by qPCR. The qPCR results consisted with the microarray results, indicating a high confidence level of our data (Fig. 1C). To further reveal the DE profile of lncRNAs and mRNAs upon CVB3 infection, we employed hierarchical clustering analysis. The results showed that the differentially expressed lncRNAs and mRNAs perfectly distinguished CVB3-infected cells from uninfected control cells (Fig. 1D, 1E).
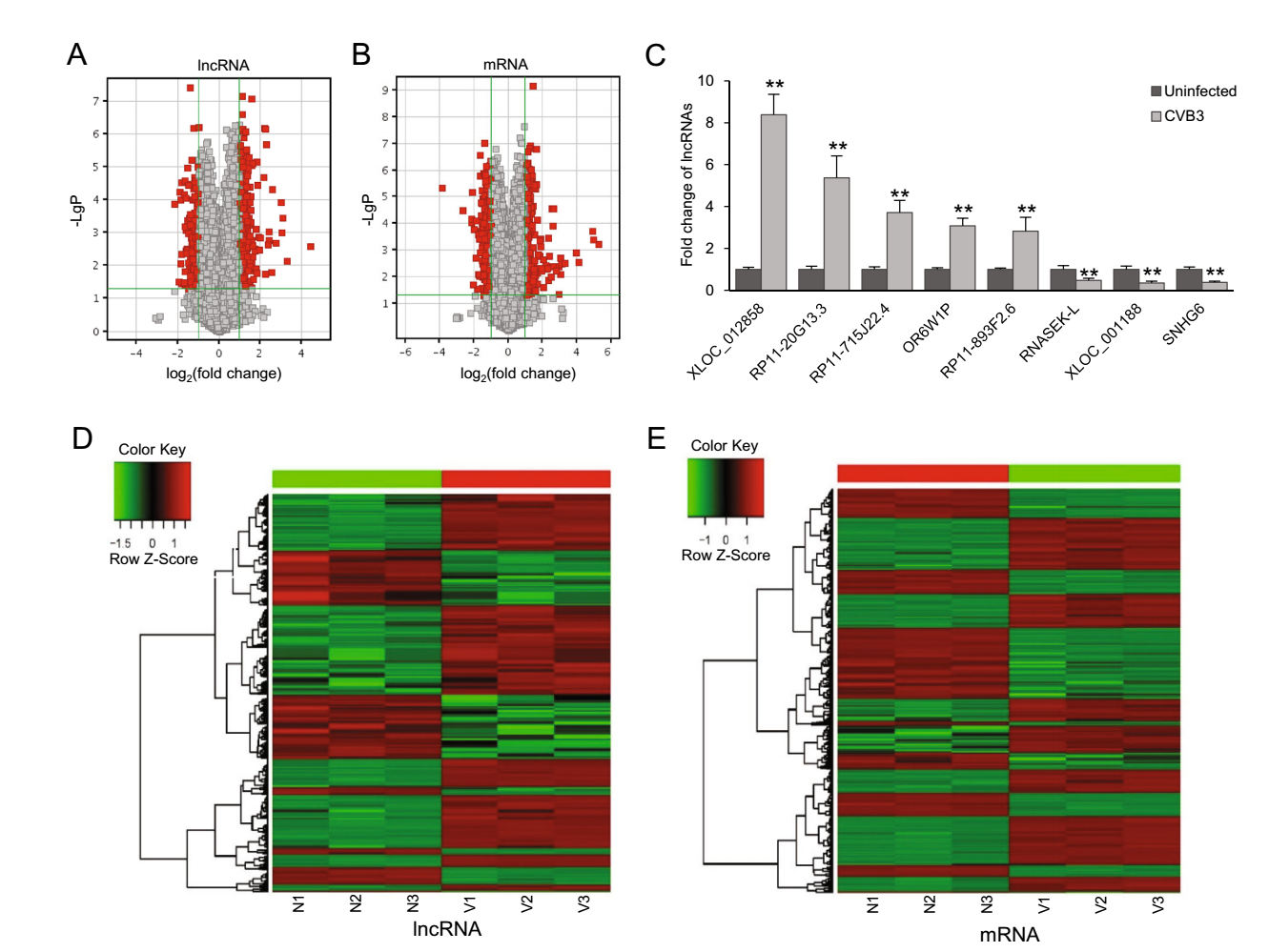
Figure 1. Differentially expressed lncRNAs and mRNAs shown in microarray and qPCR verification. Volcano plots are used to present the changes of lncRNAs (A) and mRNAs (B) in CVB3-infected cells. The vertical axis corresponds to the negative logarithm of p value with base 10 (-LgP) and the horizontal axis represents the logarithm of fold change with base 2 (log2(fold change)). The red points represent the differentially expressed lncRNAs or mRNAs with statistical significance (P < 0.05). C qPCR verification for the expression of five up-regulated and three down-regulated lncRNAs in uninfected and CVB3-infected cells. GAPDH served as an endogenous control and the results were presented as the fold change of the corresponding uninfected control (**P < 0.01). D lncRNA hierarchical clustering and E mRNA hierarchical clustering. Red color indicates up-regulation and green color indicates down-regulation. Every column represents a tissue sample and every row represents an lncRNA/mRNA probe. V represents CVB3-infected cells and N represents uninfected control cells.
-
In order to demonstrate the altered signaling pathways involving the differentially expressed mRNAs, we performed KEGG pathway analyses. The significance of a pathway was evaluated by an index of enrichment (IE) equal to "-log10 (P-value)". Therefore, the larger IE and the smaller P-value indicated the higher significant level of the pathway alteration. We counted the number of differentially expressed mRNAs in each pathway groups. According to the results, the most up-regulated pathways included asthma, rheumatoid arthritis, antigen processing and presentation, allograft rejection, and graft-versus-host disease (Fig. 2A). The down-regulated pathways included glutathione metabolism, arginine and proline metabolism, platinum drug resistance, non-alcoholic fatty liver disease, and drug metabolism–cytochrome P450 (Fig. 2B).
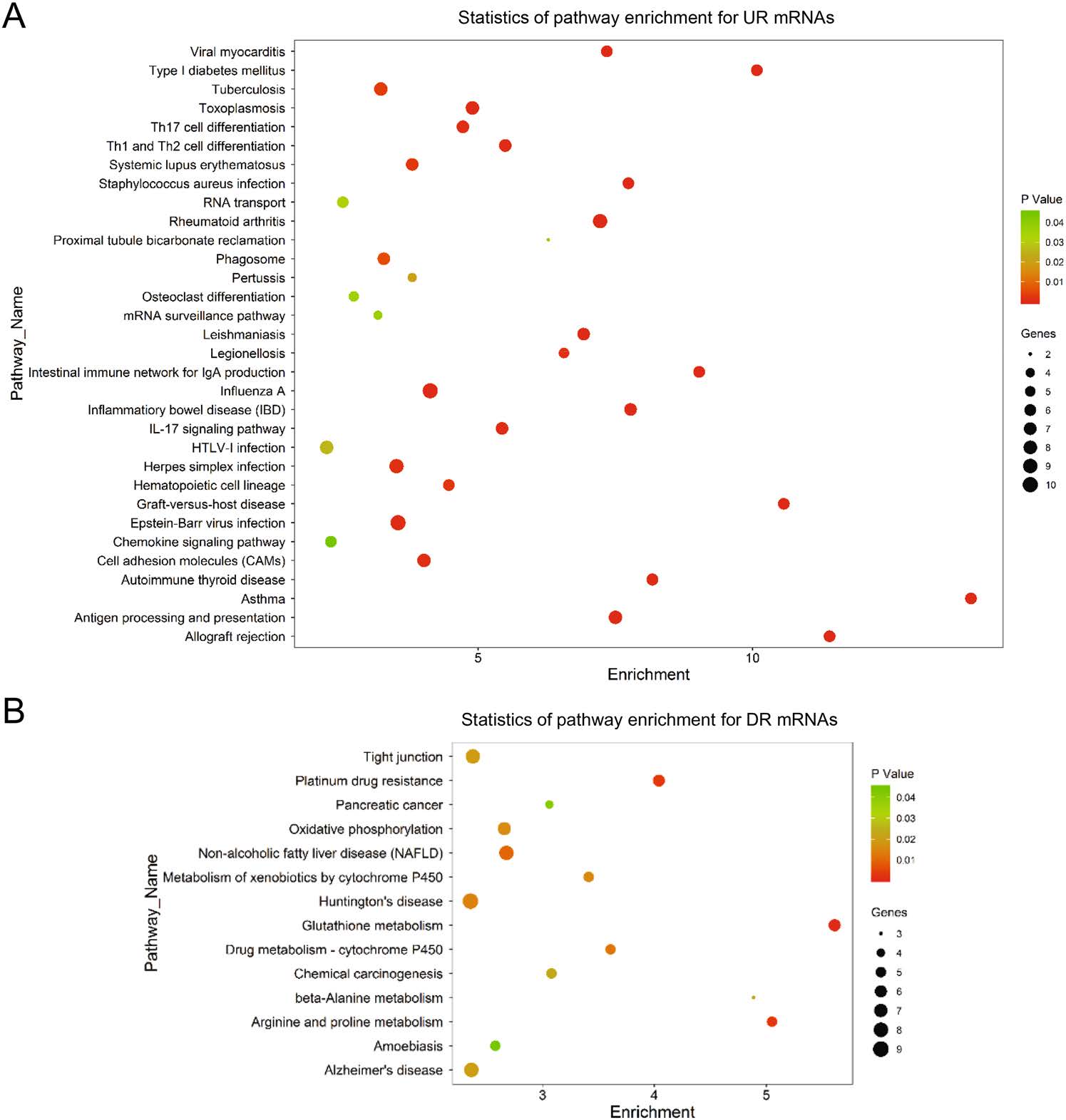
Figure 2. Pathway analysis of differentially expressed mRNAs in CVB3 infected cells. A total of 665 differentially expressed mRNAs were chosen in pathway analysis. The dot plots present the enrichment of these mRNAs in every pathway. The color of each dot corresponds to the P-value which indicates the significant level of change of each pathway. The size of each dot show the number of mRNAs involved in the corresponding pathway. The horizontal axis represents the enrichment level of the pathways. A higher enrichment level means larger change in CVB3 infection when the P-values are the same. A Pathway enrichment for upregulated (UR) mRNAs. B Pathway enrichment for down-regulated (DR) mRNAs.
-
Correlation network is an innovative tool for analyzing the correlation between lncRNAs and mRNAs. According to these correlations among mRNAs and lncRNAs, we were able to discover (ⅰ) the lncRNA affecting mRNA expression, (ⅱ) the lncRNAs which play central roles within the network and (ⅲ) the new functional mechanisms of lncRNAs can be discovered. To build the correlation network of lncRNA-mRNA expression, we first calculated the significant level of correlation for each lncRNA-mRNA pair and totally selected 3125 pairs with P < 6 × 10-6, including 203 lncRNAs and 401 mRNAs (Supplementary Table S4 and Figure S1). In the correlation network, the degree of a certain lncRNA or mRNA meant the number of mRNAs or lncRNAs, respectively, within the network correlating with it, which denoted the centrality. The lncRNAs and mRNAs with highest Correlation degrees were listed in Fig. 3, respectively. On average, each lncRNA was correlated with 15.4 mRNAs, while each mRNA was correlated with only 7.89 lncRNAs. Among the up-regulated lncRNAs, RP11-1021N1.2 was the one with the highest mRNA correlation degree, and for the downregulated lncRNAs, SNHG11 was the top one regarding the mRNA Correlation degree. As for the mRNA, TMED9 was correlated with the most lncRNAs. Based on the correlation profile, we built a correlation network of lncRNAmRNA expression (Supplementary Figure S1).
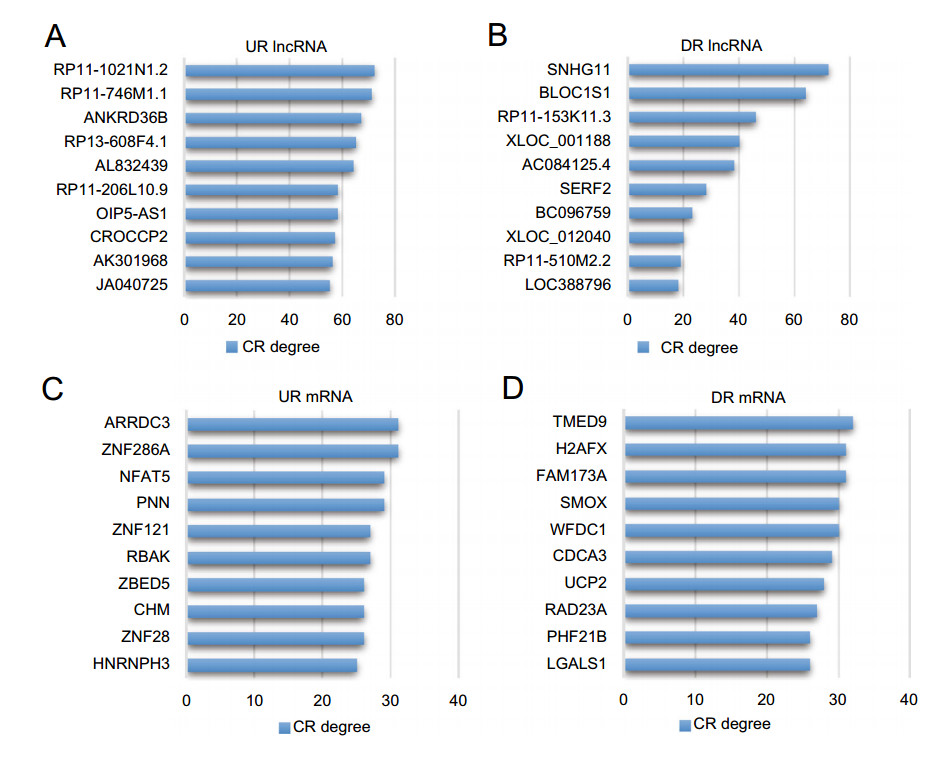
Figure 3. The top correlated lncRNA-mRNA pairs. Correlation levels of 203 lncRNAs and 401 mRNAs were evaluated by correlation degrees (CR degrees) which is equal to counts of significantly correlated lncRNA-mRNA pairs (P < 6 × 10-6) involving the corresponding lncRNA or mRNA. A Up-regulated (UR) and B down-regulated (DR) lncRNAs with top 10 CR degrees. C UR and D DR mRNAs with top 10 CR degrees.
-
To verify the reliability of the correlation network, we selected the mRNA of NFAT5, a protein significantly inhibiting CVB3 infection (Qiu et al. 2016), as the central mRNA to build a sub-network of lncRNA-mRNA correlation. In this sub-network, 27 lncRNAs positively correlated to NFAT5 expression were up-regulated in CVB3 infected cells while 3 lncRNAs negatively correlated to NFAT5 expression were down-regulated (Fig. 4A), according with the up-regulation of NFAT5 mRNA shown in our mRNA expression profile.
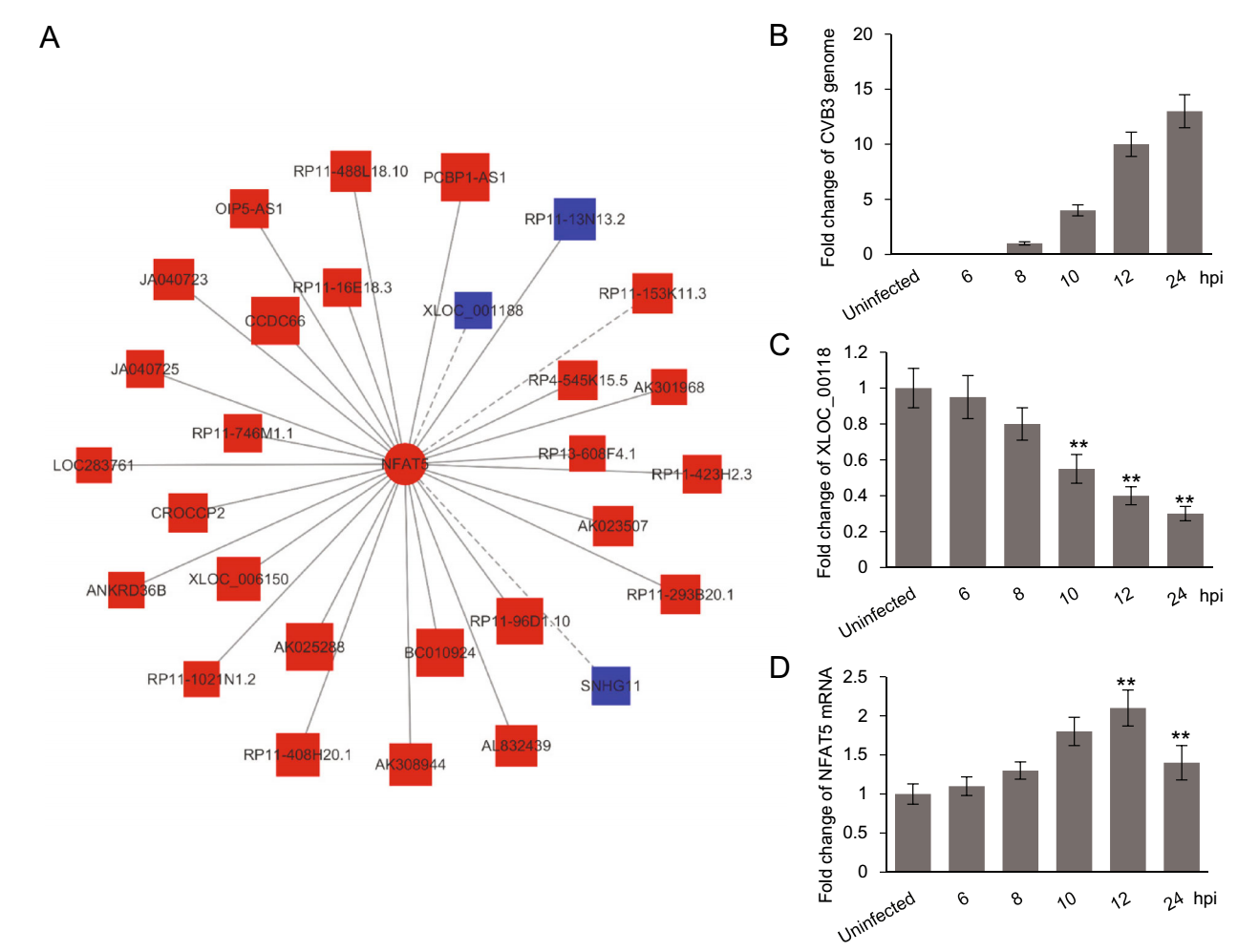
Figure 4. The correlation between lncRNA XLOC_001188 and NFAT5 mRNA. A The lncRNAs correlated to NFAT5 mRNA. The red color indicates up-regulation while the blue color indicates down-regulation. The solid lines mean positive correlation and the dashed line mean negative correlation. B–D HeLa cells were infected with CVB3 at 0.5 MOI. At indicated time points, the RNA was extracted from the cells and subjected to qPCR analysis for the levels of CVB3 genomic RNA (B), XLOC_001188 (C) and NFAT5 mRNA (D). Every time point was repeated for three times. The results were normalized to the corresponding mRNA levels of GAPDH, presented as the fold change to the 8 hpi sample (CVB3 genome) or uninfected group (XLOC_00118 and NFAT5 mRNA), and then subjected to student t test (**P < 0.01).
Among the lncRNAs correlated with NFAT5, XLOC_001188 was a most down-regulated one and was preliminarily verified by qPCR (Fig. 1C). Therefore, we selected XLOC_001188 and NFAT5 as a representative lncRNA-mRNA pair to do further verification. HeLa cells were infected with CVB3 of 0.5 MOI. At 6, 8, 10, 12 and 24 h post infection (hpi), the cells, together with the uninfected control, were subjected to RNA extraction and qPCR quantification for the levels of XLOC_001188, NFAT5 mRNA and CVB3 genomic RNA. As shown in Fig. 4B, the genomic RNA of CVB3 was observed at 8 hpi and increased at the later time points, while the decrease of XLOC_001188 started at the same time point (Fig. 4C), coinciding with the increase of NFAT5 mRNA (Fig. 4D). Nevertheless, the NFAT5 mRNA was decreased at 24 hpi, probably due to the severe cell death at the late point of viral infection. These results indicate a negative correlation between XLOC_001188 and NFAT5 mRNA in a timedependent pattern during CVB3 infection, which is consistent with the prediction. Similar results were also observed on NALCN-AS1 and NALCN mRNA (data not shown), together supporting the reliability of the lncRNAmRNA correlation network.
To further elucidate the relationship between XLOC_001188 and NFAT5 and its role in CVB3 infection, we extracted the sequence of XLOC-001188 (Fig. 5A) from a database of human large intergenic noncoding RNAs (Cabili et al. 2011). Based on the sequence, a specific siRNA was designed to knock down XLOC-001188 expression in CVB3-infected HeLa cells. Then, we detected the levels of NFAT5 mRNA and CVB3 genomic RNA. Upon siRNA challenge, we observed a dramatic decrease of XLOC-001188 (Fig. 5B), sustaining the efficiency of the siRNA we used. When XLOC-001188 was knocked down, the level of NFAT5 mRNA (Fig. 5C) was significantly increased and CVB3 genomic RNA was reduced (Fig. 5D), supporting our hypothesis that XLOC-001188 is negatively correlated with NFAT5 and NFAT5-mediated antiviral property.
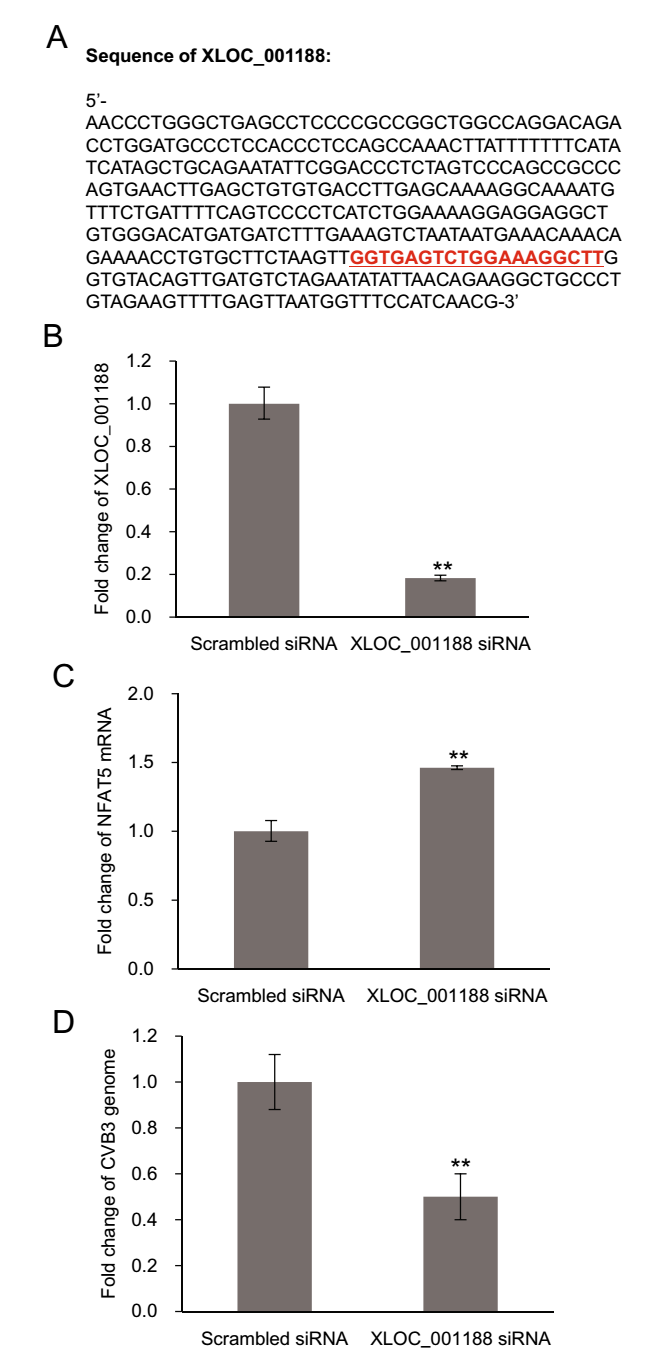
Figure 5. The negative correlation between lncRNA XLOC_001188 and NFAT5 mRNA and its influence on CVB3 replication. A Partial sequence of lncRNA XLOC_001188. The red underlined sequence is the target sequence of the XLOC_001188 siRNA. HeLa cells infected by CVB3 at 0.5 MOI for 10 h. HeLa cells were transfected with XLOC_001188 siRNA or scrambled control siRNAs, and then were subjected to CVB3 infection at 0.5 MOI for 10 h. The RNA levels of XLOC_001188 (B), NFAT5 mRNA (C), and CVB3 genome (D) were detected by qPCR. All RNA levels were normalized to the level of GAPDH mRNA. Every time point had three duplicates and the results are displayed as mean ± SD (**P < 0.01).
-
Based on the profiles of lncRNA-mRNA correlation and KEGG pathways of the differentially expressed mRNAs, we predicted the potential pathways related to the differentially expressed lncRNAs (Supplementary Table S5). Similar to the correlation network above, the correlation degrees of lncRNAs and pathways were calculated by the number of correlated counterparts linked (Supplementary Table S6). According to the correlation degree, the most significantly up-regulated pathways included those related to mRNA processing, viral infection and immune response (Fig. 6A), while the top-ranked down-regulated pathways included those related to protein turnover and pro-survival signals (Fig. 6B). RP11-1023L17.1 and RP11-145F16.2 were the up-regulated lncRNAs with the highest pathway correlation degree (Fig. 7A). Interestingly, SNHG11, the lncRNA with the top mRNA correlation degree, also had the highest pathway correlation degree among all downregulated lncRNAs, indicating a central role in the regulatory network (Fig. 7B). To investigate the molecular correlations within the pathways in detail, we selected RP11-145F16.2, RP11-1023L17.1 and SNHG11, as well as RP11-1021N1.2, the lncRNA with the top mRNA degree, as the core lncRNAs to establish a sub-network of correlation (Fig. 7C). According to the sub-network, we found that SNHG11 and RP11-1021N1.2 showed a perfect overlap on their correlated mRNAs that these two top-degree lncRNAs are correlated with the same mRNAs but in opposite directions, indicating a strong correlationship between these two lncRNAs in the regulatory network. Also, RP11-1023L17.1 was partially embedded in the SNHG11-RP11-1021N1.2 network by sharing several common correlated mRNAs such as ARRDC3, BRE, CCRN4L, CDCA3, hnRNPH3 and ZNF295. RP11- 145F16.2 was comparatively isolated but was still linked to the above network by sharing correlated mRNAs with RP11-1023L17.1, including HLA-DQA1, NEMF, IRF2BP1, FIBP, TIMP1 and MTX1. These results indicated potential synergistic regulation or mutual regulation of these lncRNAs on gene expression.
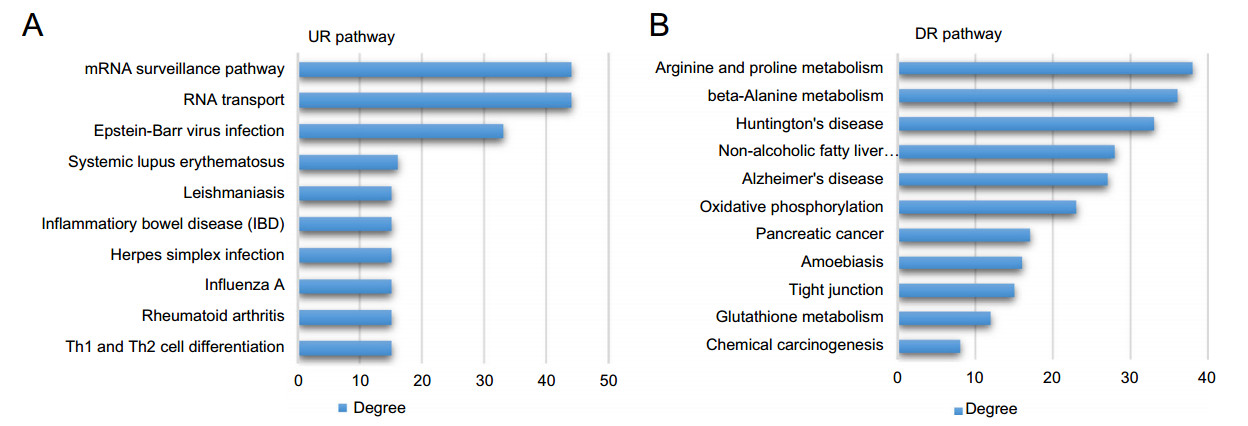
Figure 6. The pathways correlated to differentially expressed lncRNAs. The correlation between a pathway and a lncRNA was established based on their common mRNAs. The correlation levels of pathways to lncRNAs were evaluated by correlation degrees (CR degrees) which is equal to counts of lncRNAs correlated. A Up-regulated (UR) pathways with top 10 CR degrees. B Down-regulated (DR) pathways with top 10 CR degrees.
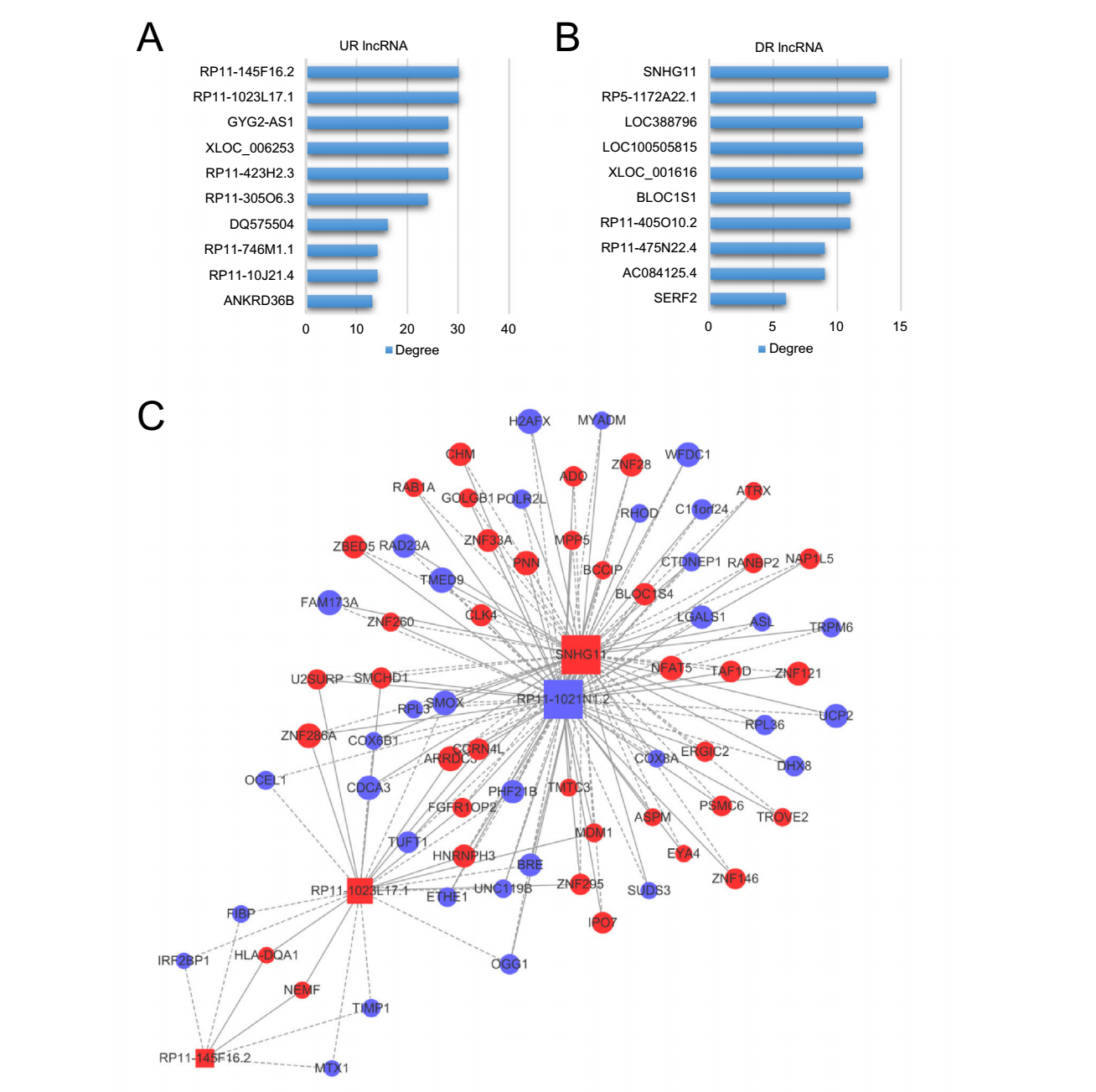
Figure 7. Pathway analysis of differentially expressed lncRNAs in CVB3 infection. The significance of lncRNAs regarding pathways were evaluated with correlation degrees (CR degrees) which is equal to counts of pathways correlated. A Up-regulated (UR) lncRNAs with top 10 CR degrees. B Down-regulated (DR) lncRNAs with top 10 CR degrees. C The lncRNA-mRNA correlation network derived from four core lncRNAs, RP11-145F16.2, RP11-1023L17.1, SNHG11 and RP11-1021N1.2.
LncRNA and mRNA Expression Profile in CVB3 Infection
Pathway Analysis for Differentially Expressed mRNAs
Correlation Network of LncRNA-mRNA Expression
Verification of the Correlation between lncRNAs and NFAT5 mRNA
Pathway Analysis of the Differentially Expressed lncRNAs
-
LncRNAs are an important subgroup of non-coding RNAs and actively transcribed in large numbers in human genome (Carninci et al. 2005; Birney et al. 2007). Attributing to the rapid development of RNA-seq in the past decade, more than 27, 000 lncRNAs have been identified in human and at least 27% of transcription across the human genome is believed to produce lncRNAs (Kapranov et al. 2007; Hon et al. 2017). LncRNAs are widely involved in various physiological and pathological processes, including viral infection. The sophisticated interplay between lncRNAs and viruses is likely to provide novel biomarkers or drug targets for diagnosis or treatment of viral infectious diseases. However, the role of lncRNAs in picornavirus infection has barely been reported. Therefore, in this study, we used CVB3, a typical picornavirus, as a model to provide a first glance of lncRNA involvement in picornavirus infection.
LncRNA-mRNA integrated microarrays screened 700 differentially expressed lncRNAs in CVB3-infected human cells compared to uninfected controls, most of which have not been described in current lncRNA database (Amaral et al. 2011; Quek et al. 2015). Unlike microRNAs, the mode of action of lncRNA has not been systematically demonstrated. Previous reports indicated that lncRNAs function through various mechanisms such as chromatin remodeling (Li et al. 2013; Orom and Shiekhattar 2013; Lam et al. 2014; Ounzain and Pedrazzini 2015), microRNA sponges (Ebert et al. 2007), ceRNAs (Karreth et al. 2011; Tay et al. 2011; Kallen et al. 2013) and antisense transcripts (Katayama et al. 2005). In fact, usually the functional mechanism of lncRNAs in a certain scenario could be quite different from others. Thus, it is hard to establish a bioinformatic program, which is widely used in microRNA study to predict the targets and function of lncRNAs. To circumvent this dilemma, we established a correlation network to pair differentially expressed lncRNAs and differentially expressed mRNAs in our study based on their expression profiles. In this way, we could indirectly predict the function of lncRNAs according to the mRNA-function analysis.
Our functional study revealed a high consistency of pathways involving differentially expressed lncRNAs and differentially expressed mRNAs. Both up-regulated lncRNAs and mRNAs are mainly involved in cellular pathways related to nucleic acid processing, immune response and inflammation, while down-regulated lncRNAs and mRNAs are mainly involved in pathways related to protein degradation and pro-survival signals. This consistency implies potential synergetic action of lncRNA and mRNA, which may be direct interaction, mutual regulation or signaling transduction. The overlap of overall functional profile well supports our strategy which predicts lncRNA function based on the lncRNA-mRNA correlation. In addition, the pathway alterations revealed by differentially expressed lncRNAs and mRNAs also match our current knowledge about CVB3 infection. For instance, CVB3 infection up-regulates RNA protective proteins to stabilize its own genomic RNA (Qiu et al. 2016). CVB3- induced up-regulation of pro-inflammatory factors is one of the major causes of inflammatory diseases (Fung et al. 2016). Also, the protein degradation via autophagy is significantly inhibited during CVB3 infection (Wong et al. 2008), which leads to protein overload and activates unfolded protein response (Zhang et al. 2010). As a cytolytic virus, it is widely accepted that CVB3 induces apoptosis to favor its viral progeny release (Yuan et al. 2003; Zhang et al. 2010). These facts sustain the reliability of our functional analysis.
According to the correlation network and pathway analysis of lncRNAs and mRNAs, lncRNAs generally have higher correlation degrees than mRNAs, indicating that lncRNA may have larger interacting range and play more regulatory roles. Considering the much lower abundance of lncRNA than that of mRNAs in cells (Ravasi et al. 2006; Cabili et al. 2011) and the high specificity of lncRNAs in different tissues (Cabili et al. 2011) or developmental stages (Yan et al. 2013), we reasonably speculate that lncRNAs play regulatory roles on multiple signaling pathways rather than execute housekeeping functions like most proteins coded by mRNAs. Therefore, lncRNAs are probably biomarkers or drug targets with more specificity and effectivity than proteins.
One lncRNA-mRNA correlation verified in our study is the pair of XLOC_001188 and NFAT5 mRNA. We have showed in a previous study that NFAT5 plays critical roles in inhibiting CVB3 replication (Qiu et al. 2017). In that study, we observed the increase of NFAT5 mRNA during CVB3 infection but could hardly explain the phenomenon. Here, according to the lncRNA-mRNA correlation network, we have found and preliminarily verified that XLOC_001188, a lncRNA down-regulated in CVB3 infection, is negatively correlated with NFAT5 mRNA, which could potentially explain the mechanism underlying the increase of NFAT5 mRNA in CVB3 infection. Considering the anti-viral activity of NFAT5, the XLOC_001188-NFAT5 co-regulation might be a host cell defense against CVB3 infection, which is an interesting direction for the future study of lncRNAs' roles in CVB3 infection.
In the correlation and pathway analysis of lncRNAs, we found four lncRNAs with the highest Correlation degrees for mRNAs or pathways, including SNHG11, RP11- 1023L17.1, RP11-145F16.2 and RP11-1021N1.2, none of which were previously reported in viral infection. SNHG11 holds the highest level for both correlation degrees for mRNAs and pathways among all down-regulated lncRNAs, implying critical position in the regulatory network. SNHG11 is a small nucleolar RNA (snoRNA) positively regulating cell proliferation. It is true that cell proliferation is slowed down during CVB3 infection (Luo et al. 2003), but the direct interaction between CVB3 and SNHG11 is quite unlikely, since the intracellular life cycle of CVB3 is exclusively located in the cytoplasm and viral components can barely enter the nucleus (Garmaroudi et al. 2015). Thus, CVB3 probably interplays with SNHG11 via an intermediate protein.
Interestingly, SNHG11 and RP11-1021N1.2 were correlated to the same group of mRNAs exactly, but each mRNA was oppositely correlated to the two lncRNAs. Since SNHG11 was down-regulated in CVB3 infection while RP11-1021N1.2 was up-regulated, thus the alterations of these two lncRNAs caused the same change of mRNAs during CVB3 infection. Considering the extraordinary coincidence of SNHG11 and RP11- 1021N1.2, it is reasonable to speculate that one lncRNA was probably upstream to the other one in a regulatory pathway. This result indicates a potential mutual regulation between the two lncRNAs which was barely reported in previous studies, though more evidence is needed to support this speculation.
RP11-145F16.2, RP11-1023L17.1 and RP11-1021N1.2 were significantly up-regulated during CVB3 infection, but their functions remain obscure to date. Our correlation network has revealed several interesting genes correlated to these lncRNAs. For instance, BRE mRNA was negatively correlated to RP11-1023L17.1 and RP11-1021N1.2, indicating an up-regulation of BRE in CVB3 infection. BRE protein was reported to promote the degradation of p53, a classical tumor-suppressor protein (Jin et al. 2017). Interestingly, CVB3 infection indeed induces degradation of p53 which benefits CVB3 replication (Gao et al. 2010). These findings imply that RP11-1023L17.1 and RP11- 1021N1.2 may be involved in the CVB3-p53 mutual regulation. In addition, IRF2BP1 was negatively correlated to RP11-145F16.2 and RP11-1023L17.1, indicating a downregulation of IRF2BP1 in CVB3 infection. IRF2BP1 protein is a transcriptional corepressor binding to the C-terminal repression domain of IRF2, a negative regulator of many interferon (IFN)-responsive genes (Childs and Goodbourn 2003). IFN response is a critical antiviral pathway. Thus, down-regulation of IRF2BP1 may be a cellular protective response against CVB3 infection in which RP11-1023L17.1 and RP11-1021N1.2 are involved.
In conclusion, the strategy utilizing lncRNA-mRNA correlation is reliable to predict lncRNA function in CVB3 infection, providing a useful tool for the lncRNA studies. By this approach, we have revealed a negative correlation between the lncRNA XLOC_001188 and the antiviral gene NFAT5. In addition, we have also identified four critical lncRNAs, SNHG11, RP11-145F16.2, RP11-1023L17.1 and RP11-1021N1.2, which potentially affect CVB3 replication. Although the results are quite prelimnary and more verification is required, this is the first study indicating the involvement of lncRNAs in CVB3 infection. These findings bring a new perspective for enterovirus studies, which may provide potential drug targets for therapy development against CVB3 infection.
-
This work was supported by the National Natural Science Foundation of China (81101234 to Lei Tong; 81571999, 81871652 to Zhaohua Zhong; 31470260 to Xingyi Ge; 81672007 to Wenran Zhao; 81772188 to Yan Wang), the Foundation of Heilongjiang Provincial Postdoctor of China (LBH-Z11076 to Lei Tong), the China Postdoctoral Science Foundation (2015M580269 to Lexun Lin), the Research Foundation of Education Bureau of Heilongjiang Province (12511176 to Lei Tong), the Hu-Xiang Youth Talents Scholar Program of Hunan Province (2017RS3017 to Xingyi Ge), Health and Family Planning Commission of Heilongjiang Province (2016-165 to Lexun Lin), the Provincial Natural Science Foundation of Hunan Province (Grant Number 2019JJ50035 to Ye Qiu) and the Fundamental Research Funds for the Central Universities of China (Grant Number 531107051162 to Ye Qiu). We are grateful to the technical support from Heilongjiang Provincial Key Laboratory of Pathogens and Immunity and Heilongjiang Provincial Science and Technology Innovation Team in Higher Education Institutes for Infection and Immunity of Harbin Medical University. We thank Jing Li (Cnkingbio Company Ltd, Beijing, China) for technical support.
-
LT, YQ, XG and ZZ designed the experiments. HW, YQ, LL, YW, WX, WZ and HH carried out the experiments. LT, YQ, YZ, GZ, MHZ and DY analyzed the data. YQ wrote the paper. LT, YQ, DY, XG and ZZ checked and finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: