HTML
-
Viruses are the one of the most ancient life forms, and causative agents of approximately 70% diseases of human and animals. Studies related to the invasion and reproduction mechanism of viruses provide significant insight into the virus life cycle, which can be later exploited for the development of drugs and therapeutics against viral diseases (Simmonds and Domingo 2011). Viral infections in cells are visualized by immunofluorescence which requires the infected cell to be fixed using paraformaldehyde, therefore immunoflorescence is not a preferred/suitable method for tracking and studying viral movement in live cells (Simmonds and Domingo 2011). Single virus tracking techniques enables better understanding of various stages of viral infection in live cells, thereby allowing a better understanding of viral infection process, including attachment, entry, transport, replication and release. Till date, this technology has been successfully used to elucidate the infection pathway of many viruses (Liu et al. 2012; Zhang et al. 2013b; Hou et al. 2019; Nathan and Daniel 2019).
In order to track the virus in a live cells, viruses and cellular constituents need to be labeled using fluorophore indicators. These fluorophore indicators have been classified into three categories: organic dyes (e.g., Cy5), fluorescent proteins (e.g., GFP) and inorganic nanoparticles (e.g., QDs). Organic dyes are very small in size and diverse kinds have been widely used in single particle tracking (Zhang et al. 2013a; Sun et al. 2017). However, their fluorescence is not strong and the signal fades quickly during in situ and real-time monitoring (Liu et al. 2016). Fluorescent protein genes can be fused with the viral or cellular protein genes as one open reading frame (ORF). Similar to organic dyes, the fluorescent proteins also possess weak fluorescence stability, making it difficult to track the interaction of virus and cellular constituents over a long period of time (Zhang et al. 2013a; Sun et al. 2017).
Quantum dots (QDs) are novel inorganic semiconducting nanomaterials, having unique optical properties such as high brightness, superior photostability and large stokes shift (Wen et al. 2017; Zheng et al. 2017). They have been proven to be ideal fluorescent materials for labeling and tracking single viruses (Liu et al. 2016). Recent studies, utilizing QDs labelled viruses provided specific details about active interactions between viruses and cellular components (Lakadamyali et al. 2003; Joo et al. 2011; Huang et al. 2012; Zheng and Wang 2013). After labeling with streptavidin (SA), alkynyl or succinimide on their surface, the viruses can be linked with the QDs (Liu et al. 2016; Sun et al. 2017; Wen et al. 2017; Zheng et al. 2017). The SA–QDs system has been used to study several large viruses, such as influenza virus, ectromelia virus, pseudorabies virus and Baculovirus (Huang et al. 2012; Zhang et al. 2013a; Wang et al. 2016; Sun et al. 2017).
The porcine reproductive and respiratory syndrome virus (PRRSV) is a positive-stranded RNA enveloped virus with mature virion size ranges from 40 to 60 nm and belongs to the family Arteriviridae. It causes severe economic losses to swine industry worldwide (Lunney et al. 2016; Jiang et al. 2015; Conzelmann et al. 1993; Nelsen et al. 1999; Mu et al. 2013; Bulgakov et al. 2014; Stankevicius et al. 2016). Despite causing severe economic losses worldwide, not much is known about PRRSV infection pathway.
In this study, we labeled the PRRSV with QDs by streptavidin–biotin affinity system and dissected its entry pathway and movement pattern in live cells. We studied the interactions between PRRSV and cell structural elements, mainly microtubules and microfilaments, along with two receptors: non-muscle myosin heavy chain Ⅱ-A (NMHC Ⅱ-A) and vimentin. This work will give a good example for the application of QDs on the smaller virus tracking. At the end of the study we were able to provide some novel and important insights into PRRSV infection mechanism.
-
Streptavidin-modified quantum dots (QDs–SA) were purchased from Wuhan Jiayuan quantum dots Co., LTD (China). Their emission wavelength was 605 nm. Marc- 145 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco) supplemented with 2% (v/v) fetal bovine serum (Gibco) at 37 ℃ in 5% CO2 condition. Highly pathogenic porcine reproductive and respiratory syndrome virus (H-PRRSV) was used in this study.
-
Marc-145 cells were infected with H-PRRSV at a multiplicity of infection (MOI) of 1. The supernatants were collected 48 h after transfection, and cell debris was removed by centrifugation at 4000 × g for 20 min. Virions were collected by filtering the supernatants through 0.45 mm filter (Millipore) and subsequently centrifuged at 25, 000 rpm at 4 ℃ for 90 min (Beckman SW28 rotor) on 5 mL of a 30% (w/v) sucrose cushion in Tris–EDTA buffer (TE, 0.01 mol/L Tris, pH 7.4, 1.0 mmol/L EDTA). The virion pellets were resuspended in TE buffer.
To biotinylate the virus, the concentrated virions and Sulfo-NHS-LC-Biotin (Thermo ScientificTM) were incubated for 1–2 h with continuous shaking at 37 ℃. Unbound Sulfo-NHS-LC-Biotin was removed by passing the reaction mixture through the NAP-5 column (GE Healthcare Illustra), and the biotinylated viruses were stored at 4 ℃. For confocal microscopy, the Marc-145 cell was adsorbed with biotinylated virus (4 ℃ for 90 min), in which condition PRRSV only can attach the cell surface but not enter the cell. After adsorption cells were washed with PBS buffer, QDs–SA were added into the petridishes and incubated for 20 min at 4 ℃. After washing the cells 3 times with PBS buffer, the cells were observed using confocal microscope.
-
Based on the sequences of NMHC Ⅱ-A and vimentin of Cercopithecusaethiops in GeneBank, the genes were synthesized by Sangon Biotech Co., Ltd (Shanghai). After confirming the sequences, the genes were cloned into pEGFP-N1 plasmid digested with EcoRI/SacⅡ and NheⅠ/ HindⅢ respectively.
-
Marc-145 cells were transfected with EGFP-Lifeact plasmid for labeling microfilaments (actin filament), GFPMAP4 (microtubule-associated-protein 4) plasmid for labeling microtubule, pEGFP-N1-NMHC Ⅱ-A plasmid for labeling non-muscle myosin heavy chain Ⅱ-A and pEGFPN1-vimentin for labeling vimentin.
Briefly, the Marc-145 cells were grown to 70% confluence in 12 well tissue culture plates and transfected with a reaction mixture consisting of 3 μL lipofectamine reagent (Invitrogen) along with 50 μL Opti-MEM Ⅰ reduced serum medium (Gibco) and 1.5 μg DNA making a final volume of 100 μL. Lipofectamine-DNA mixture was incubated at room temperature for 30 min, and later added to the cell culture. Media was changed after incubation for 6 h, and the transfected cells were cultured for a total of 24 h. The transfected cells were dispersed with trypsin digestion and used for confocal microscopy.
-
In order to disrupt the microfilaments and microtubules, the Marc-145 cells were dispersed in Tyrode's plus buffer supplemented with 20 mmol/L cytochalasin D (Cyto D) (Sigma) and Tyrode's plus buffer with 60 mmol/L nocodazole (Sigma), respectively for 60 min. Cells were washed three times with PBS buffer and infected with the biotinylated virus for 30 min at 4 ℃. The infected cells were washed again with PBS buffer prior to confocal microscopy observation.
-
Marc-145 cells were infected with biotinylated viruses. After washing with PBS buffer, cells were fixed in 4% (w/v) paraformaldehyde for 20 min at room temperature. 0.1% Triton-100 was added to improve the permeability of virus envelope and cytomembrane. Cells were washed again with PBS, and incubated with 5% (w/v) BSA in PBS for blocking for 30 min. The cells were then incubated with monoclonal primary antibody (Veterinary Medical Research & Development, 1:400 diluted) against nucleocapsid (N protein) of PRRSV at 37 ℃ for 2 h. The cells were then washed extensively with PBS containing 1% BSA and incubated with secondary antibody (Dylight 649 conjugated goat anti-mouse IgG, Thermo, 1:500 diluted) at 37 ℃ for 45 min. After washing with PBS buffer, the SA–QDs were added. Finally virus co-localization with cellular machinery was analyzed by confocal microscope.
-
The cells were imaged by a spinning-disk confocal microscope (Andor Revolution XD) equipped with an Olympus Ⅸ-81 microscope, a CO2 online culture system (INUBG2-PI), a Nipkow disk type confocal unit (CSU 22, Yokogawa) and an EMCCD (Electron-Multiplying Charge Coupled Device, AndoriXon DV885K). The images were acquired using EMCCD. For simultaneous two-color imaging, the fluorescence was imaged alternately on the EMCCD by two channels.
Co-localization of viruses with cellular components was analyzed with Imaging-Pro Plus software. The light intensity analysis was developed by Image J software. Kymograph images were processed by Andor IQ software. The trajectories were generated by software Imaging-Pro Plus software. The imaging of momentary speed variations were analyzed by Origin 8.0. Each frame in the movies was processed by a gauss filter to remove background noise.
-
Carbon-coated copper grids were put onto 50 μL virus suspension, and incubated for 5 min. After drying on filter paper, the samples were stained with 2% phosphotungstic acid and dried naturally. The grids were examined on a Hitachi H-7000 FA transmission electron microscope at 200 kV.
Quantum Dots, Cells and Viruses
Virus Infection, Purification and Labeling
Construction of the Recombinant Plasmids
Transient Transfection
Inhibition of Filaments and Microtubules
Immunofluorescence Co-localization Analysis
Fluorescence Imaging and Analysis
Transmission Electron Microscopy (TEM)
-
Our labeling strategy included 4 critical steps. Firstly, PRRSV virions were propagated in Marc-145 cells and purified using sucrose density gradient centrifugation. Secondly, purified PRRSV were biotinylated using SulfoNHS-LC-Biotin. Thirdly, the Marc-145 cells were incubated with biotinylated viruses at 4 ℃, allowing the viruses to adsorb on cell membranes but not to pass through the membranes. Finally, the cells bound by biotinylated viruses were incubated with SA–QDs to get QD labeled PRRSV (Fig. 1).
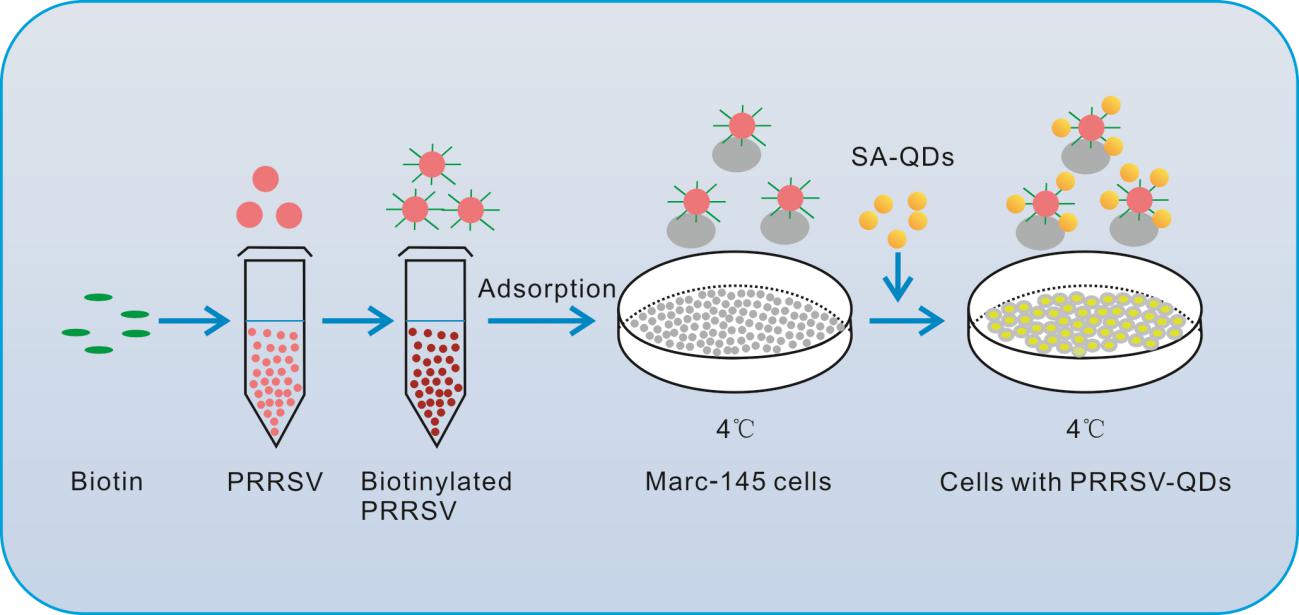
Figure 1. General labeling scheme of single-particle PRRSV with quantum dots. Marc-145 cells were absorbed by biotinylated PRRSV at 4 ℃, and then the streptavidin– quantum dots (SA–QDs) were added.
The viral ultrastructure was analyzed using transmission electron microscopy (TEM) to assess the effect of SA–QD labeling on virus shape and size. TEM analysis showed that the virions were intact with about 40 nm diameter, and no other structural irregularity was observed as a result of labelling (Fig. 2A). Next the effect of labeling on viral infectivity was analyzed using 50% tissue culture infective dose (TCID50) and one-step growth curve assay. As seen in Fig. 2B, 2C, there was only minor reduction in viral infectivity after labeling with QDs, indicating that the labeled viruses were infectious. These results demonstrated that the labeling strategy was feasible to label small viruses such as PRRSV.
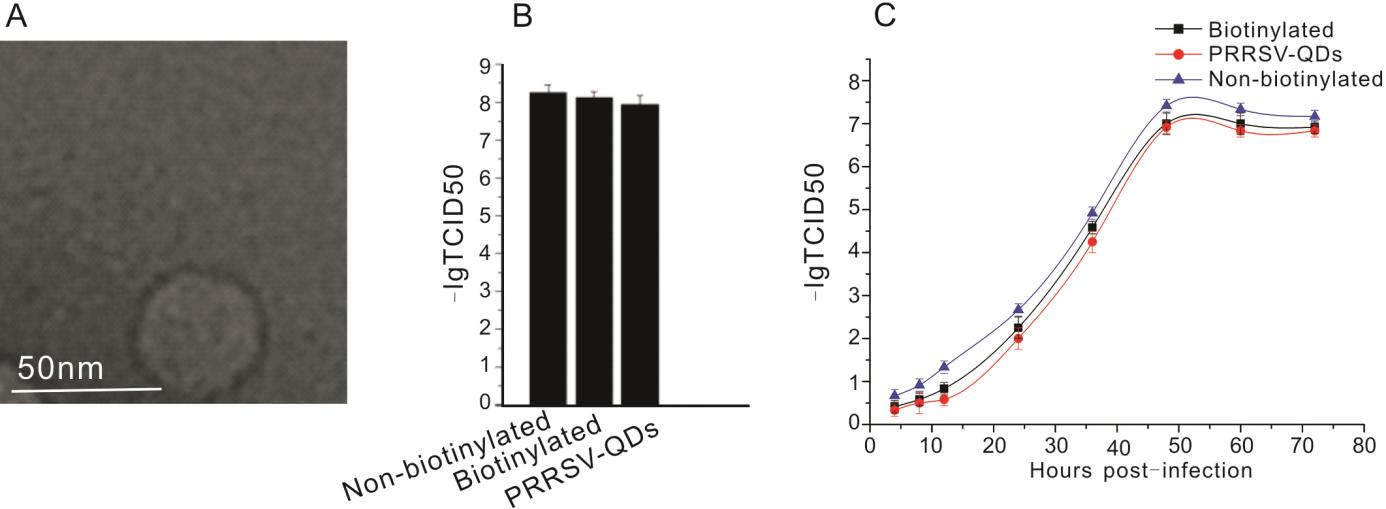
Figure 2. PRRSV virion labeling and infectivity assay. A Transmission electron microscopy (TEM) image of biotinylated PRRSV virions purified by sucrose density gradient centrifugation. B Analysis of viral infectivity by 50% tissue culture infectivity dose (TCID50) of non-biotinylated PRRSV, biotinylated PRRSV, and PRRSV-QDs. C The growth curve of non-biotinylated PRRSV, biotinylated PRRSV and PRRSV-QDs in one step replication assay. The experiment was repeated three times.
-
In order to evaluate the labeling specificity, PRRSV-QDs were added to Marc-145 cells for attachment at 4 ℃. As shown in Fig. 3A, Marc-145 cells infected by PRRSV-QDs showed strong and discrete fluorescence on the cell surface (Fig. 3A a–c). In the negative control groups (Marc-145 infected by non-biotinylated PRRSV, Marc-145 infected by biotinylated PRRSV, Marc-145 infected by non-biotinylated PRRSV and incubated with SA–QDs) (Fig. 3A d–f), weak fluorescence signals were detected. These results proved that PRRSV was successfully labeled with SA–QDs, which was a prerequisite for virus singleparticle tracking.
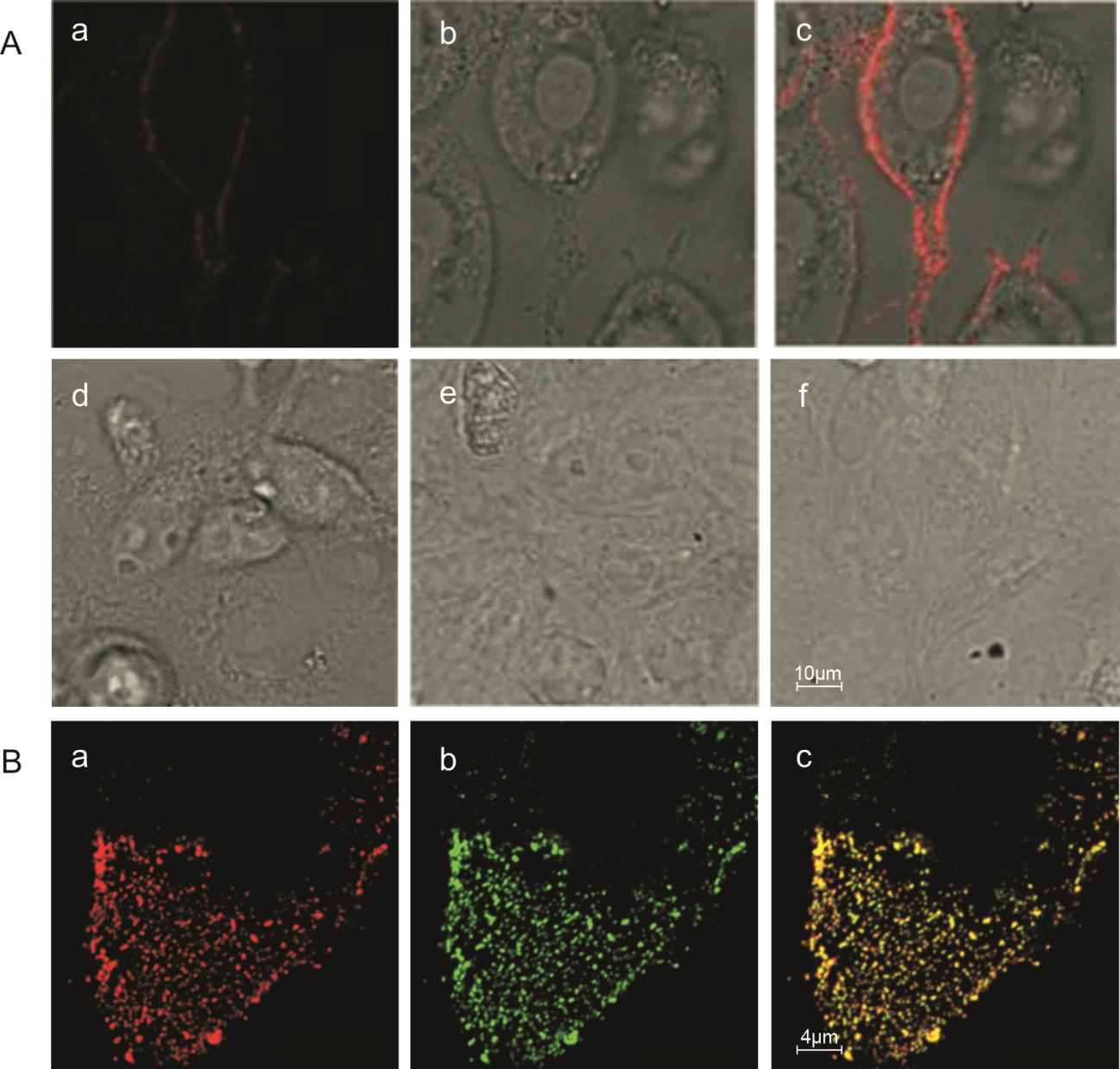
Figure 3. Specificity and efficiency of quantum dots (QDs) labeling of PRRSV. A: Specificity of quantum dots (QDs) labeling of PRRSV. A a–c: Laser field, bright-field and merge of Marc-145 cells infected with PRRSV-QDs; A d–f: negative controls of PRRSV-QDs infection. Marc-145 infected by non-biotinylated PRRSV (d), biotinylated PRRSV (e), non-biotinylated PRRSV (f) and incubated with SA– QDs, respectively. Efficiency of quantum dots (QDs) labeling of PRRSV. B a: Fluorescence of QDs of PRRSV-QDs (red), B b: Immunofluorescence of N protein of PRRSV-QDs (green), B c: The merge of B a and B b (yellow).
Co-localization analysis showed that the QDs-labeled viruses exhibited discrete fluorescent dots in the cells (Fig. 3B a). Over 98% of discrete fluorescence in the QDs image co-localized with the nucleocapsid protein (N protein) of PRRSV (Fig. 3B b, c), illustrating that the QDs signals were adequate to exhibit the presence of virus particles in live cells.
-
To investigate the interaction of PRRSV with cytomembrane, Marc-145 cell membranes were stained with Dio, a lipophilic fluorescent dye, and incubated with the biotinylated viruses for 30 min at 4 ℃. After binding of SA–QDs to the biotinylated viruses, the virions were monitored using confocal microscopy. Interestingly, we found that prior to the entry into the cell via an endosome, the viruses first oscillated on the cytomembrane (Fig. 4A, Supplementary Movie S1). The oscillatory movement of PRRSV could be due scanning for appropriate receptors for attachment and entry.
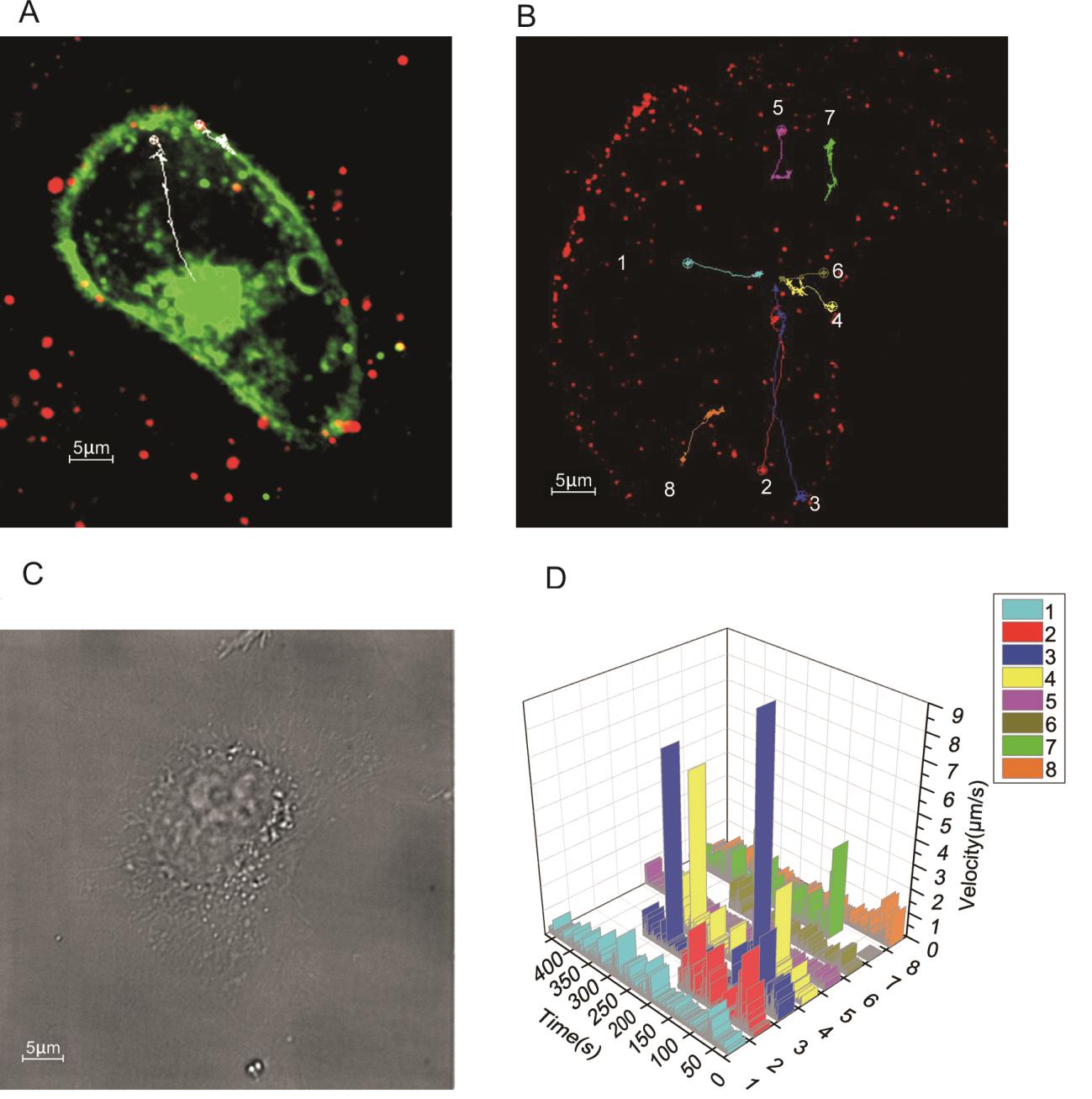
Figure 4. Tracking of PRRSV entry. A Fluorescent images of PRRSV-QDs and cytomembrane labeled with a lipophilic fluorescent dye Dio, B eight trajectories of PRRSV movement in the cell, C differential interference contrast (DIC) image of B, D velocity vs time plots of eight virions tracked during the entry process.
Viral movement and entry were tracked and imaged by monitoring multiple virions simultaneously for about 30 min with a frame interval of 500 ms, exposure time of 300 ms. It was observed that the virus particles vibrated on the membrane for just few seconds and then appeared in the cytoplasm. In the cytoplasm the viruses made fast and unidirectional movement and finally accumulated in a special region inside the cell (Supplementary Movie S2). Trajectories of 8 individual virions also demonstrated that the viruses were converging to the perinuclear region (Fig. 4B, 4C).
Based on the analysis of velocity, most virions exhibited a slow–fast–slow movement pattern and accumulated at the perinuclear region (Fig. 4D), indicating that the virus movement may be utilizing different cytoskeletal components such as microfilaments, microtubules and intermediate filaments simultaneously. The fastest instantaneous speed and average speed of intracellular transport were 7.6 μm/s and 0.22 μm/s, respectively.
-
Marc-145 cells were transfected with a GFP-MAP4 (microtubule-associated-protein 4) plasmid to label the microtubules. The movement of PRRSV-QDs on microtubules was observed using confocal microscopy. It was observed that few virions moved along the microtubules, whereas few others moved for a short distance and then disappeared. Their transport followed a typical pattern of movement: sometimes rapidly, sometimes slowly and sometimes stopping (Supplementary Movie S3). Single virus trajectory on microtubules was also recorded, and representative images were shown in Fig. 5A.
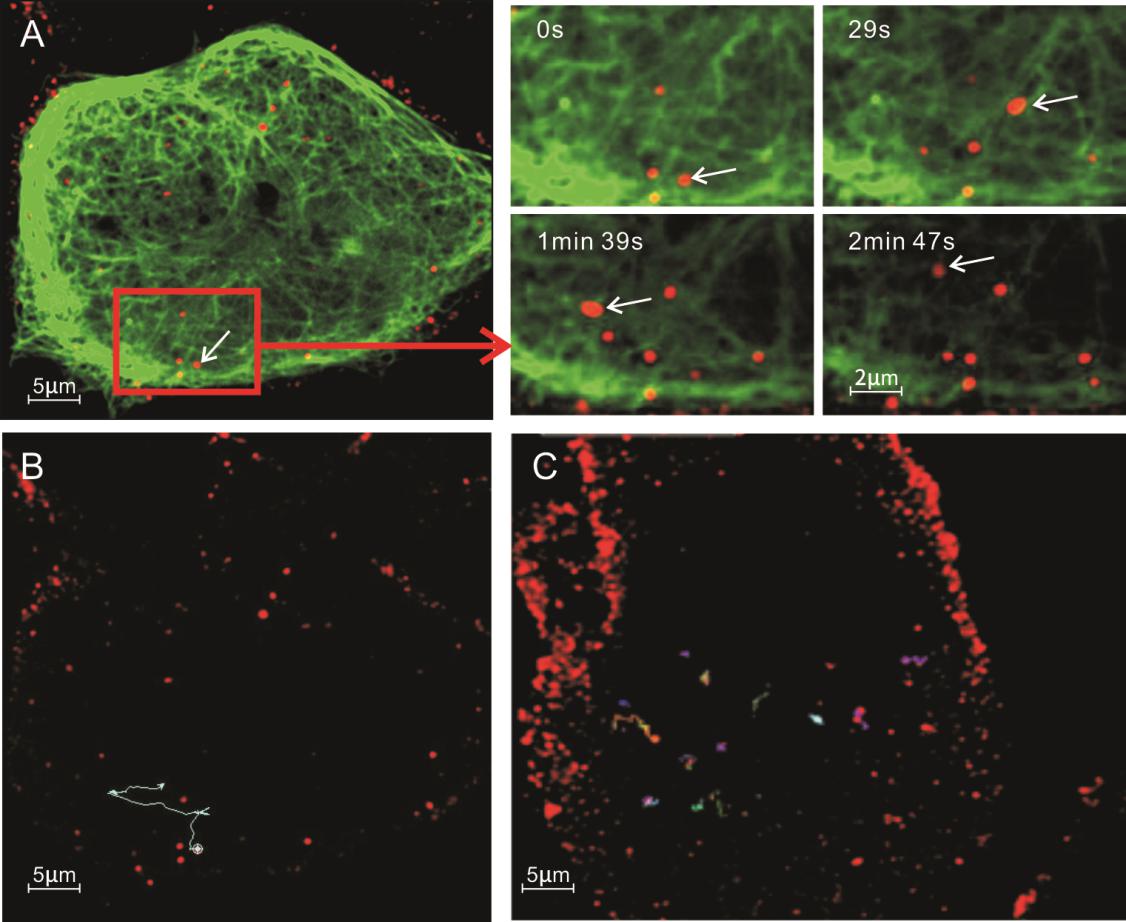
Figure 5. Interaction of PRRSVQDs with microtubules in Marc- 145 cells. A Real-time tracking of PRRSV-QDs transported along the microtubules to the perinuclear region. The right part shows single PRRSV-QDs moving on one microtubule at different time points. Microtubules are labeled with GFP. B Trajectory of individual PRRSV-QDs in living cells. C Trajectory of individual PRRSV-QDs in living cells after treatment with 60 μmol/L nocodazole.
To further confirm whether the PRRSV movement toward microtubule organizing center (MTOC) was microtubule-dependent, nocodazole, a drug to disturb microtubules, was used to treat Marc-145 cells before infection. The trajectory and velocity of the single virus particle were analyzed. In untreated cells, PRRSV-QD moved from the cell periphery to the cytoplasm with a long trajectory and showed an active movement pattern (Fig. 5B). In contrast, in nocodazole-pretreated Marc-145 cells, the viruses just vibrated rapidly in a small range (Fig. 5C). These two different transport modes indicated that nocodazole could strongly inhibit virus traffic during the course of infection. These results indicated that the PRRSV movement within the cells was microtubuledependent.
-
Marc-145 cells were transiently transfected with plasmid expressing EGFP-actin. The EGFP-actin expressing cells were infected with PRRSV-QDs and then fixed with 4% paraformaldehyde. Confocal microscopy images showed the co-localization of microfilaments and PRRSV (Data not shown). In order to understand the relation of microfilaments and PRRSV further, the trajectory of single virus was tracked in live cells. The results showed that some viruses transported from cytomembrane to nuclear periphery along one microfilament, which were long trajectories. It was tracked that some viruses moved along part of one microfilament and then onto other or disappeared (Fig. 6A, and Supplementary movie S4). Interestingly, most viruses exhibited a slow–fast–slow pattern of movement.
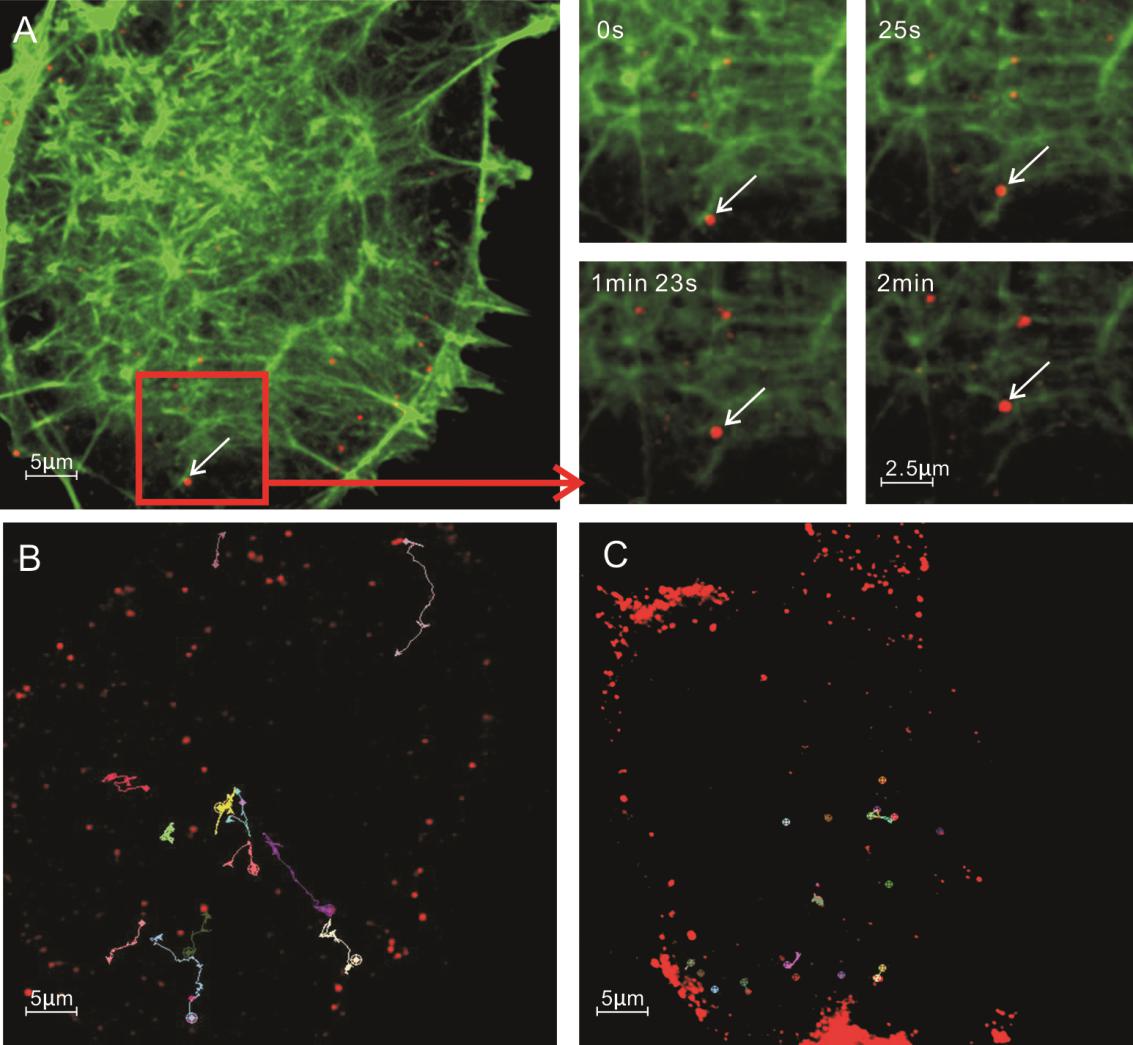
Figure 6. Microfilamentdependent entry pathway of PRRSV-QDs in Marc-145 cells. A Real-time tracking of PRRSV-QDs transported along the microfilaments to the perinuclear region. The right part shows single PRRSV-QDs moving on one microfilament at different time points. B Trajectory of individual PRRSV-QDs in living cells. C Trajectory of individual PRRSV-QDs in living cells treated with 20 μmol/L CytD.
To confirm the PRRSV transport is microfilament-dependent, CytD, a drug that disrupts microfilaments, was used to treat Marc-145 cells before infection. The trajectories of single virus from untreated to treated cells were compared. Similar to the viral movement on microtubules, disruption of microfilaments limited the range of virus movement. The virus could only move back and forth in a short distance on one microfilament (Fig. 6B, 6C).
-
Here we tracked the interaction between these two receptors and PRRSV in live cells to explore their relation in different time and space. Firstly, the two genes were synthesized based on the nucleotide sequences of vimentin and NMHC Ⅱ-A of Cercopithecusaethiops available in Genebank. The verified sequences were cloned into plasmid pEGFP-N1. When the EGFP-vimentin or EGFP-NMHC Ⅱ-A was transiently expressed in Marc-145 cells, the Vimentin and NMHC Ⅱ-A were labeled (Figs. 7A and 8A). Similar to PRRSV movement along the microtubules and microfilaments, the PRRSV could move close to the nuclear periphery using the vimentins with an average velocity of 0.19 μm/s (Fig. 7A, 7B and Supplementary Movie S5).
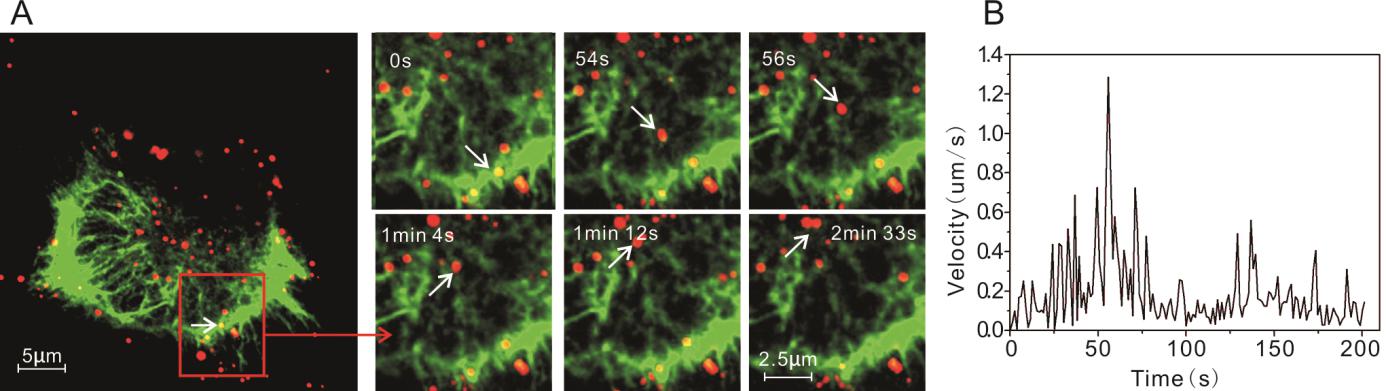
Figure 7. Vimentin-dependent entry of PRRSV-QDs in Marc-145 cells. A Real-time tracking of PRRSV-QDs transported along the vimentin to the perinuclear region. The right part shows single PRRSV-QDs moving on one vimentin at different time points. B Instantaneous speed of individual PRRSV-QDs in living cell.
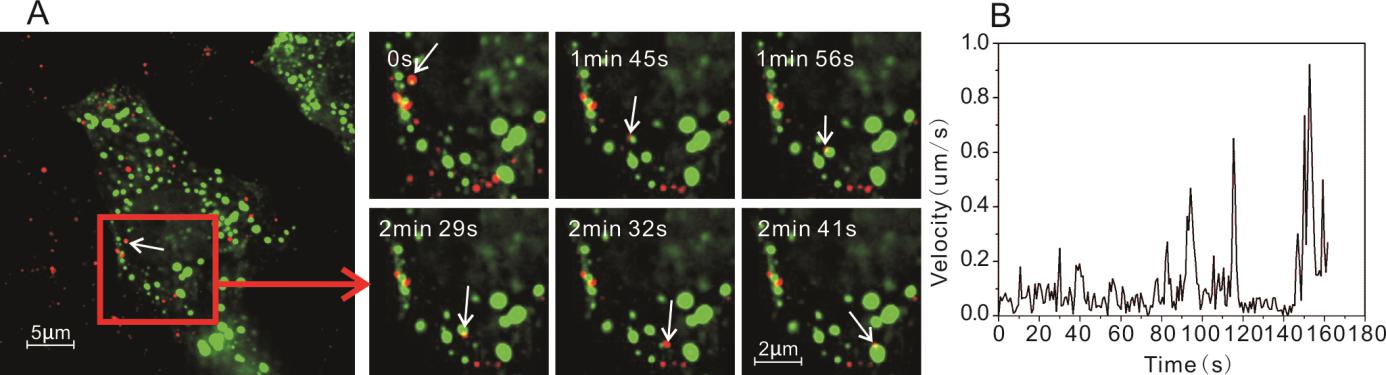
Figure 8. Interaction of PRRSV-QDs and NMHC Ⅱ-A in Marc-145 cells. A Real-time tracking of PRRSV-QDs transport and its relation with NMHC Ⅱ-A. The right part shows single PRRSV-QDs contacting with NMHC Ⅱ-A at different time points. B Instantaneous speed of individual PRRSV-QDs in living cell.
PRRSV interaction with NMHC Ⅱ-A was different in comparison to interaction with microtubules, microfilaments and vimentins. Firstly, NMHC Ⅱ-A existed in a different form from all these three proteins, which were many irregular small spheres in a free state in cytoplasm (Fig. 8A). Secondly, during the movement, the viruses made contacts with many NMHC Ⅱ-A (Fig. 8A, 8B and Supplementary Movie S6). However, more study need to be done to figure out how NMHC Ⅱ-A helps virus movement in the cell.
Biotinylation and Infectivity of PRRSV
Labeling Specificity and Efficiency
Dynamic Process of PRRSV Entry
PRRSV Endocytosis and Microtubules
Microfilament and PRRSV Endocytosis
PRRSV Infection and Two Pivotal Receptors: Vimentin and NMHC Ⅱ-A
-
PRRSV infection begins with the recognition and attachment of viral particles to receptors on the cell membrane, which are followed by the delivery of virions into the cells within vesicles via a clathrin-dependent pathway and followed by a pH-dependent fusion event (Gorp et al. 2010). However, the nature of virus and host cell receptors and attachment still remains unclear. Microtubules play key roles during movement and infectivity of several viruses (Taylor et al. 2009; Su et al. 2010; Liu et al. 2014; Dumas et al. 2015). Actins are the key protein of microfilaments. The functions of microfilaments include: muscle contraction, cell movement, intracellular transport/trafficking, maintenance of eukaryotic cell shape, and cytokinesis and cytoplasmic streaming (Bretscher 1991; Wang et al. 2015). Previous study has indicated that the cellular transport of PRRSV is facilitated by microfilaments (Kreutz and Ackermann 1996).
To date, several important receptors for PRRSV attachment, such as heparin sulfate (HS), sialoadhesin (Sn), CD163, CD151, vimentin and NMHC Ⅱ-A, have been identified. These receptors play significant roles in PRRSV infection since they are involved in virus binding, internalization or uncoating (Wang et al. 2012; Zhang and Yoo 2015; Wells et al. 2017). Of the six known receptors, the newly identified vimentin and NMHC Ⅱ-A, have been proven to be pivotal in virus infection (Wang et al. 2011; Zhang and Yoo 2015; Song et al. 2016). Vimentin plays a significant role in supporting and anchoring the position of the organelles in the cytosol (Murakami et al. 2012). Nonmuscle myosin Ⅱ is one of the main motor protein interacting with cytoskeletal actin and is involved in regulating cytokinesis, cell motility, and cell polarity in many eukaryotic cells (Chandrasekar et al. 2014). However, there is no clear understanding about the mechanism of interaction of these receptors with the virus.
In this research, we labeled the PRRSV with QDs by streptavidin–biotin affinity system. This method, with advantages of mild reaction conditions and easy operation, was appropriate to real-time tracking of the small virus in live cells. Our results showed that PRRSV vibrated on the cytomembrane first and then entered the cell within an endosome. PRRSV was transported along the microtubules, microfilament and vimentin with a slow–fast–slow movement pattern and finally accumulated in the perinuclear region. Maybe the PRRSV could transfer their paths between microtubules, microfilaments and vimentins. When viruses were transported, they contacted with many NMHC Ⅱ-A which were irregular small spheres in cytoplasm. Our data has unraveled some important features of PRRSV infection and may facilitate the application of quantum dots in real-time imaging of virus infection, including the attachment, entry, uncoating and release.
-
The authors would like to thank for the support from the National Natural Science Foundation of China (Grant Nos. 31570151 and 31490601), the Program for Science and Technology Innovation Talents in Universities of Henan Province (Grant No. 17HASTIT039), the Key Scientific Research Project of Henan Province Higher Education (16A180044) and the Open Research Fund Program of the State Key Laboratory of Virology of China (Grant No. 2017KF005).
-
XZ and ZL conceived and designed the study. ZL, PL and CW performed the experiments. ZL and DS wrote and edited the manuscript. XZ finalized the manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.


















 DownLoad:
DownLoad: