-
Dear Editor,
The family Totiviridae contains non-enveloped, double-stranded RNA viruses with genomes of approximately 4.6-7 kbp in length. These totiviruses are classified into five genera: Totivirus, Victorivirus, Trichomonasvirus, Giardiavirus, and Leishmaniavirus (Nibert 2007). They are associated with latent infections of fungal or protozoan hosts (Li et al. 2020). Recently, a few totiviruses have been identified and shown to have unexpected host distributions, including insects, plants, and even Chromalveolata (Chen et al. 2016; Koyama et al. 2015; Marchler-Bauer et al. 2017; Mor and Phelps 2016). However, most of these new viruses have not been assigned to appropriate genera by the International Committee on Taxonomy of Viruses (ICTV). Omono River virus (ORV) is an unclassified RNA virus in the family Totiviridae (Isawa et al. 2011; Okamoto et al. 2016). In this study, we reported a novel ORV isolated from a pool of Culex tritaeniorhynchus mosquitoes collected in Yunnan, China. We also updated the phylogenetic relationships of the family Totiviridae to include all genomes reported to date.
Mosquitoes were collected using ultraviolet light lamps in Yunnan, China in 2012. The captured mosquitoes were grouped according to the location and mosquito classification. Approximately 200 mosquitoes in each group were milled together and cultured with Aedes albopictus C6/36 cells. After blind-passaging by three times, the pool that caused a cytopathic effects (CPE) was further selected for RNA extraction and deep sequencing on an S5 sequencer.
We identified a new ORV from the culture products of the Culex tritaeniorhynchus pool collected in Yuanjiang County. This virus was designated as Omono River virus strain YJ according to the collection site. YJ could be stably passaged in C6/36 cells and cause a CPE (Fig. 1A-a, 1A-b). The growth curves were determined by real-time polymerase chain reaction using standard dsRNA. YJ had strong proliferative ability in C6/36 cells and reached a plateau on the third day of incubation (Fig. 1B). Transmission electron microscopy revealed that the viral particles were uniformly spherical particles approximately 25 nm in diameter (Fig. 1A-c, 1A-d). The virus particles we observed were smaller than those of previously reported viruses. (e.g., 40 nm for other ORVs and C. tritaeniorhynchus totivirus KL) (Li et al. 2020).
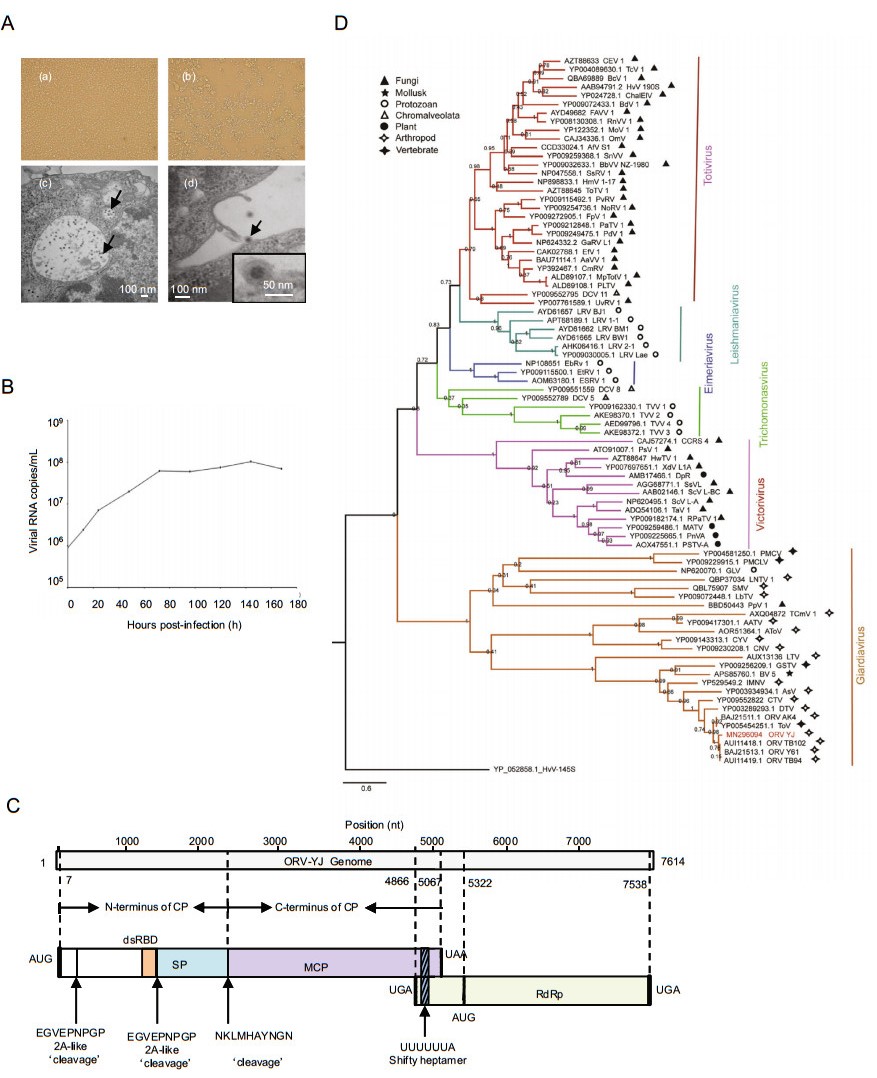
Figure 1. Isolation and characterization of this new YJ strain. A-a C6/36 cells without YJ infection; b C6/36 cells with YJ infection; c and d transmission electron microscopy images of YJ-infected C6/36 cells (arrow). B Replication kinetics of YJ in C6/36 cells. C Organization of different regions in the YJ genome. D Phylogenetic tree of RdRp protein sequences of YJ and other representative totiviruses.
The genome of YJ was 7614 nucleotides long and shared the highest identity (79% nucleotide similarity and 93.13% amino acid similarity) with the known ORV strain TB94 (GenBank Accession Number: KY264025). The complete genome of YJ was deposited in GenBank (Accession Number: MN296094). The sequence of YJ encoded two open reading frames (ORFs). ORF1 (nt 7–5067) of YJ encoded a 1687aa capsid protein, whereas ORF2 (nt 5322–7538) of YJ encoded a 739aa RNA-dependent RNA polymerase (RdRp). The two proteins were translated into fusion protein owing to a-1 ribosomal frame shift (de Lima et al. 2019). A typical heptanucleotide slippery sequence, shown as 'UUUUUUA', was found at nt positions 4896–4902 in the ORV-YJ genome. The typical heptanucleotide slippery sequence UUUUUUA was a key signal element for -1 ribosomal frame-shifting, and was found at nt positions 4896–4902 in the YJ genome (Isawa et al. 2011). Two 2A-like sequence motifs (EGVEPNPGP) were found near the N-terminus of YJ ORF1 (Dantas et al. 2016), at aa 88–96 and 502–510, respectively. These two short amino acid sequences divided the ORF1 into three products: 95-, 412-, and 1178-aa protein fragments (Fig. 1C). The 1178-aa protein was further split into 281- and 782-aa sequences, yielding NKLMHAYNGN (aa 780–789) through a different mechanism (Nibert 2007). BLASTp showed that the C-terminus of the 412-aa protein shared homology with the receptor-binding domain. However, the protein encoded by the 95- and 281-aa segments has not been elucidated.
To construct a phylogenetic tree, 82 respective RdRp protein sequences deposited before July 2019 were obtained from NCBI Taxonomy database (Supplementary Table S1). The phylogenetic tree was constructed using MEGA v7.0 with the maximum likelihood model and 1000-replicate bootstrap test. Phylogenetic analysis based on RdRp amino acid sequences revealed six monophyletic groups in the family Totiviridae (Fig. 1D), including the genera adopted by ICTV (Nibert 2007), i.e., Giardiavirus, Totivirus, Victorivirus, Leishmaniavirus, and Trichomonasvirus. Almost all unclassified viruses were grouped within one genus. Eimeria brunetti RNA virus 1 (EbRV-1), Eimeria tenella RNA virus 1 (EtRV-1), and Eimeria stiedai RNA virus 1 (EsRV-1) formed a distinguishing cluster and might be defined as a new genus, for which we propose the genus name Eimeriavirus. ORV strain YJ, as an unclassified virus in the family Totiviridae, was closely related to other ORVs and grouped in the potential Giardiavirus genus. Given the long evolutionary distance between viruses within the potential Giardiavirus genus, we believe that the clade may contain multiple genera. Further studies are required to elucidate the specific classification. The hosts of Giardiavirus are diverse, including mollusks, vertebrates, arthropods, and protozoans, suggesting that Artivirus genus may not only contain viruses that infect arthropods (de Lima et al. 2019). Viruses in other genera also exhibit host diversity, disrupting the paradigm that viruses within a group infect hosts belonging to a single kingdom. In contrast, host diversity within each genus may provide insights into the classification of groups within the family Totiviridae.
In summary, a new mosquito-specific ORV collected in Yuanjiang, China was identified. Biological and genomic analyses of YJ were also conducted. Phylogenetic analysis based on all representative species in the family Totiviridae provided a foundation for the ICTV to classify members of the family Totiviridae.
HTML
-
This research was supported by grants from the National Key R & D Program of China (2017YFC1200802), National Science and Technology Major Project of China (2018ZX10711001-003-001), the State Key Laboratories Program of China (SKLPBS1817), and the Yunnan Health Training Project of High Level Talents (D-201604).
-
The authors declare that they have no conflicts of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: